Phylogenetics and
virus evolution
Trevor Bedford (@trvrb)
Oct 24, 2019
MCB532 Human Pathogenic Viruses
Phylogeny describes evolutionary relationships
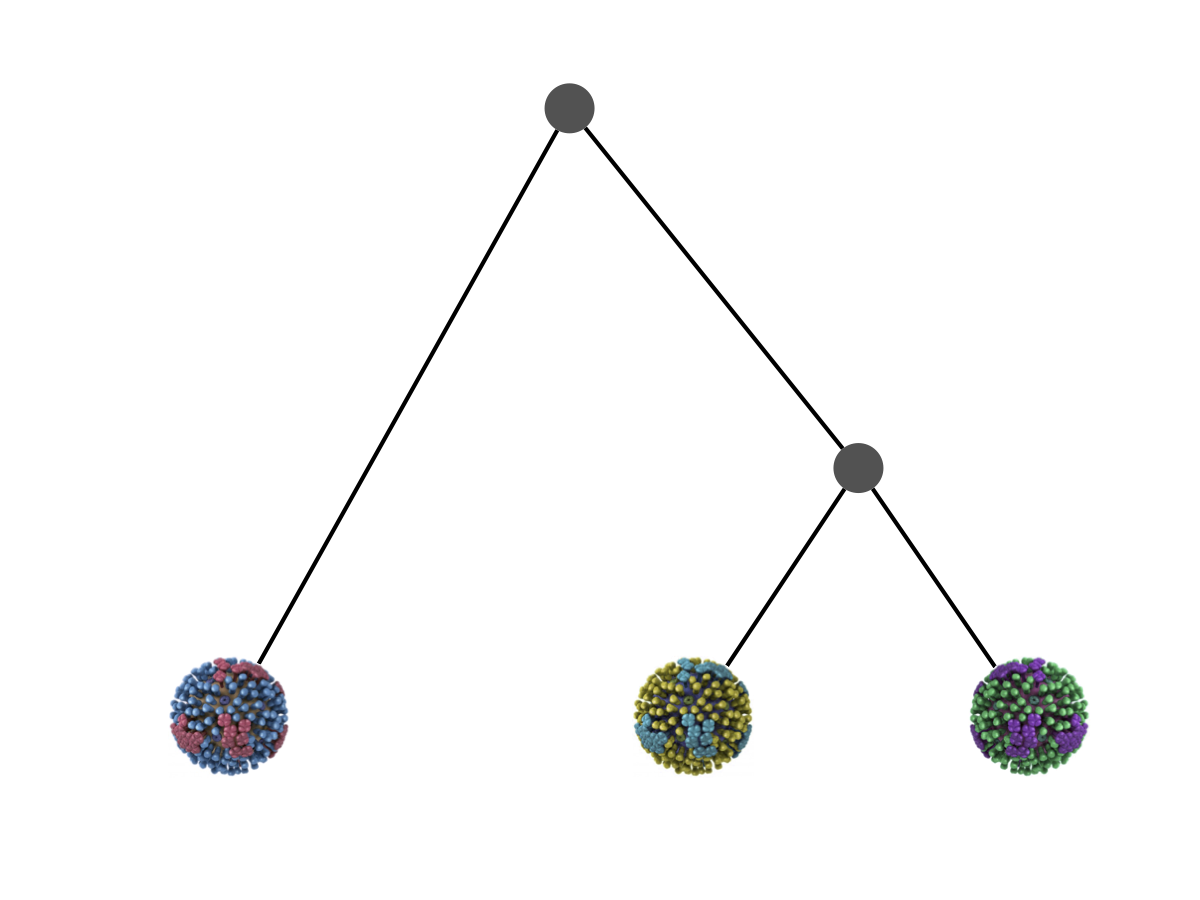
Phylogeny is usually a hypothesis based on characteristics of sampled taxa
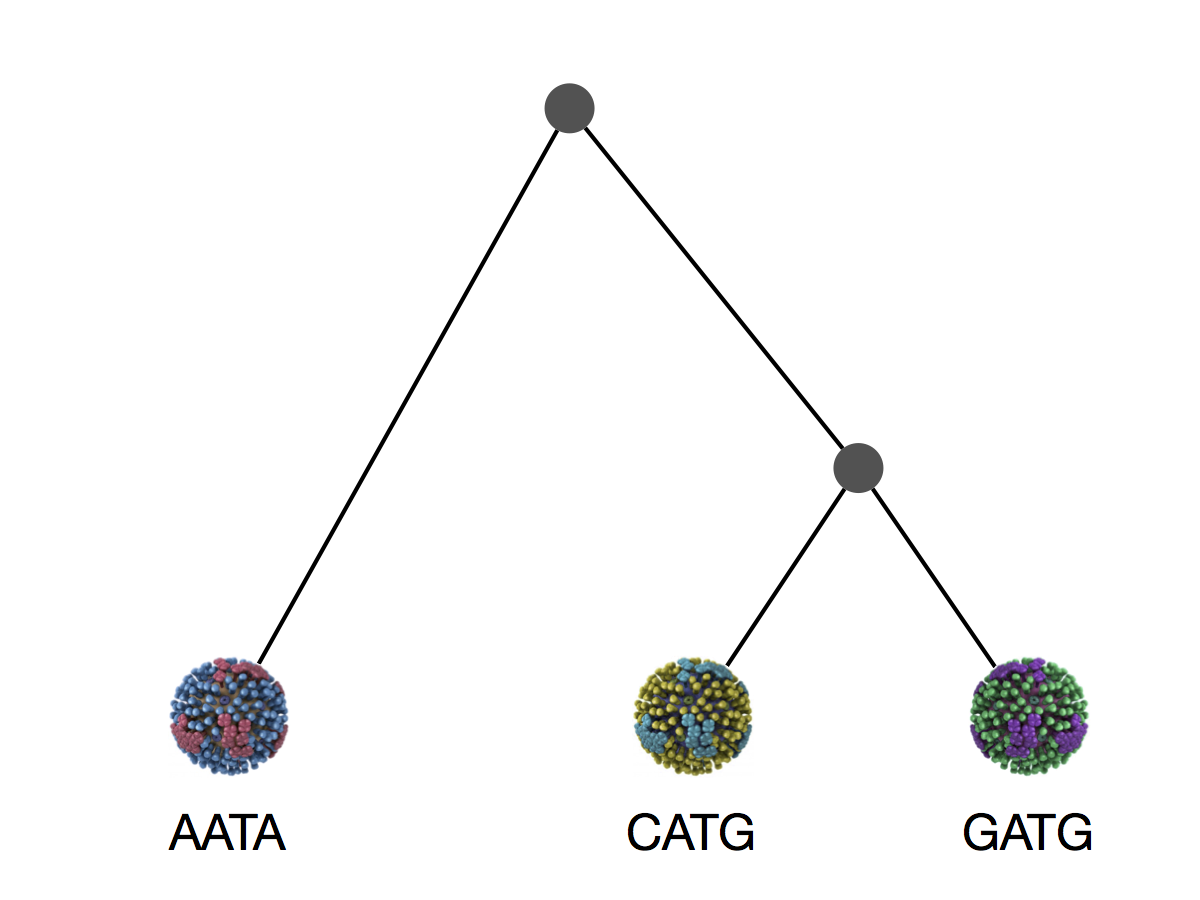
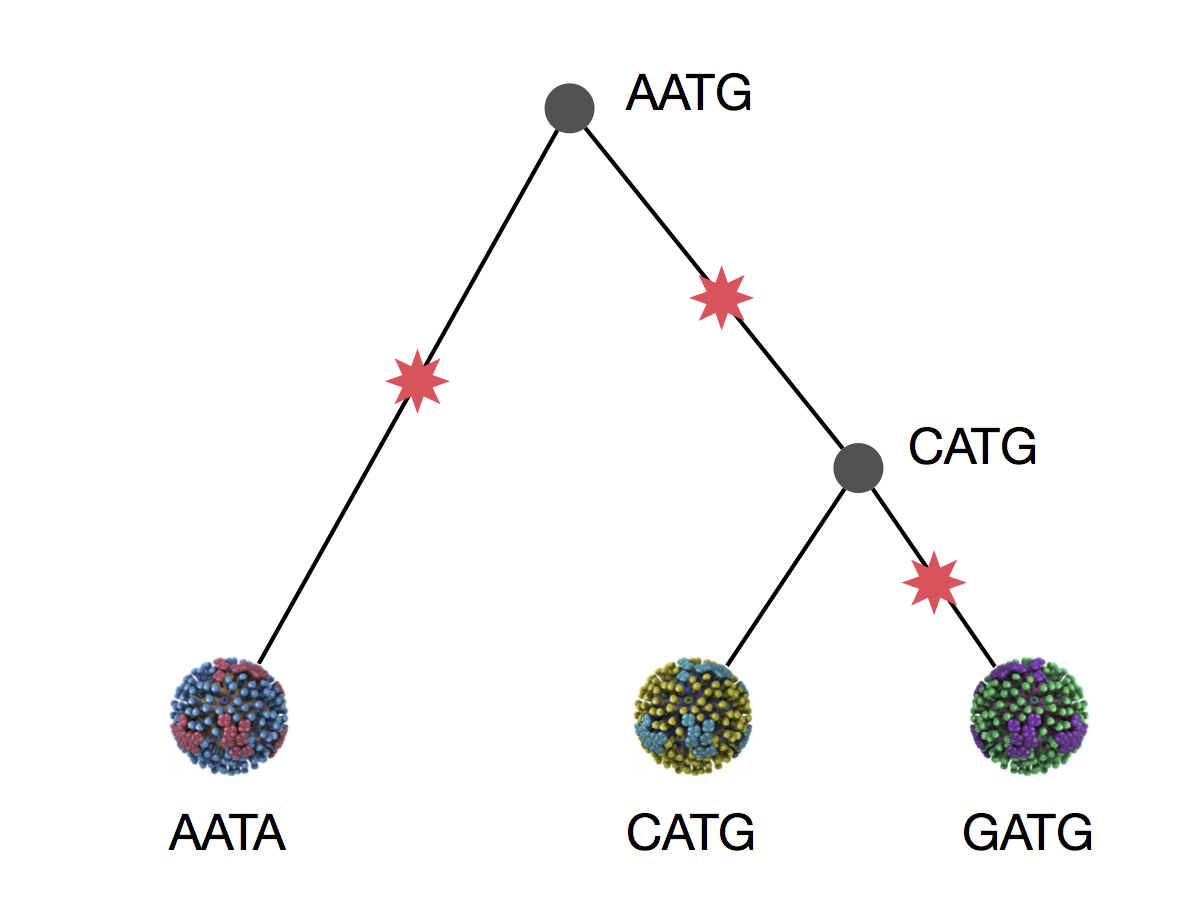
Phylogeny implies a series of mutational events leading to observed tip states
Phylogenetic inference
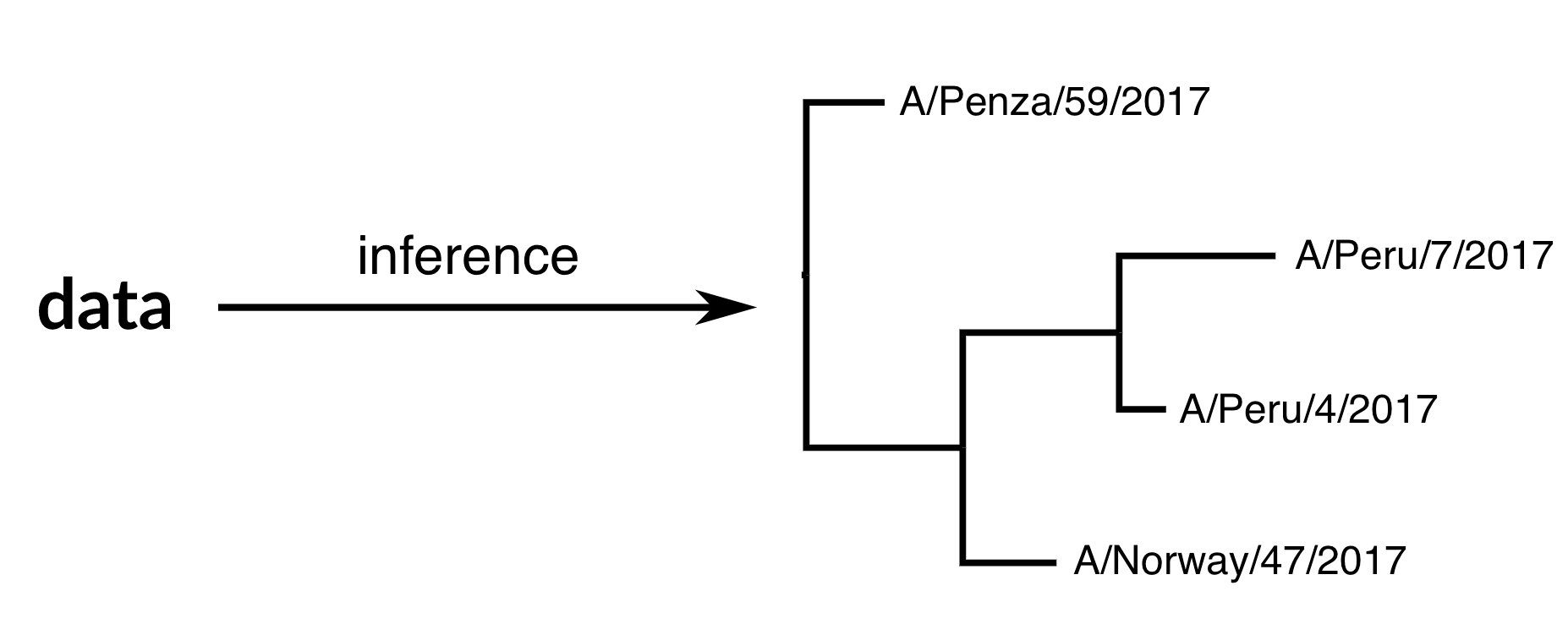
"Data" is generally a sequence alignment

Phylogeny structures site patterns
Tree space is vast
There are (2n-3)!! rooted trees for n taxa
- 3 taxa: 3 trees
- 5 taxa: 105 trees
- 10 taxa: 34,459,425 trees
- 20 taxa: 8.2 × 1021 trees
- 50 taxa: 2.8 × 1076 trees
- 100 taxa: 3.3 × 10184 trees
Solution space is rugged
Types of phylogenetic inference methods
- Distance-based (neighbor-joining, fast, heuristic)
- Parsimony (fast, "model-free")
- Maximum likelihood (infers model of mutation, accurate, examples: FastTree, RAxML)
- Bayesian (like ML, but requires prior, produces estimates of uncertainty, examples: MrBayes, BEAST)
Inference is a tree topology, branch lengths and ancestral states
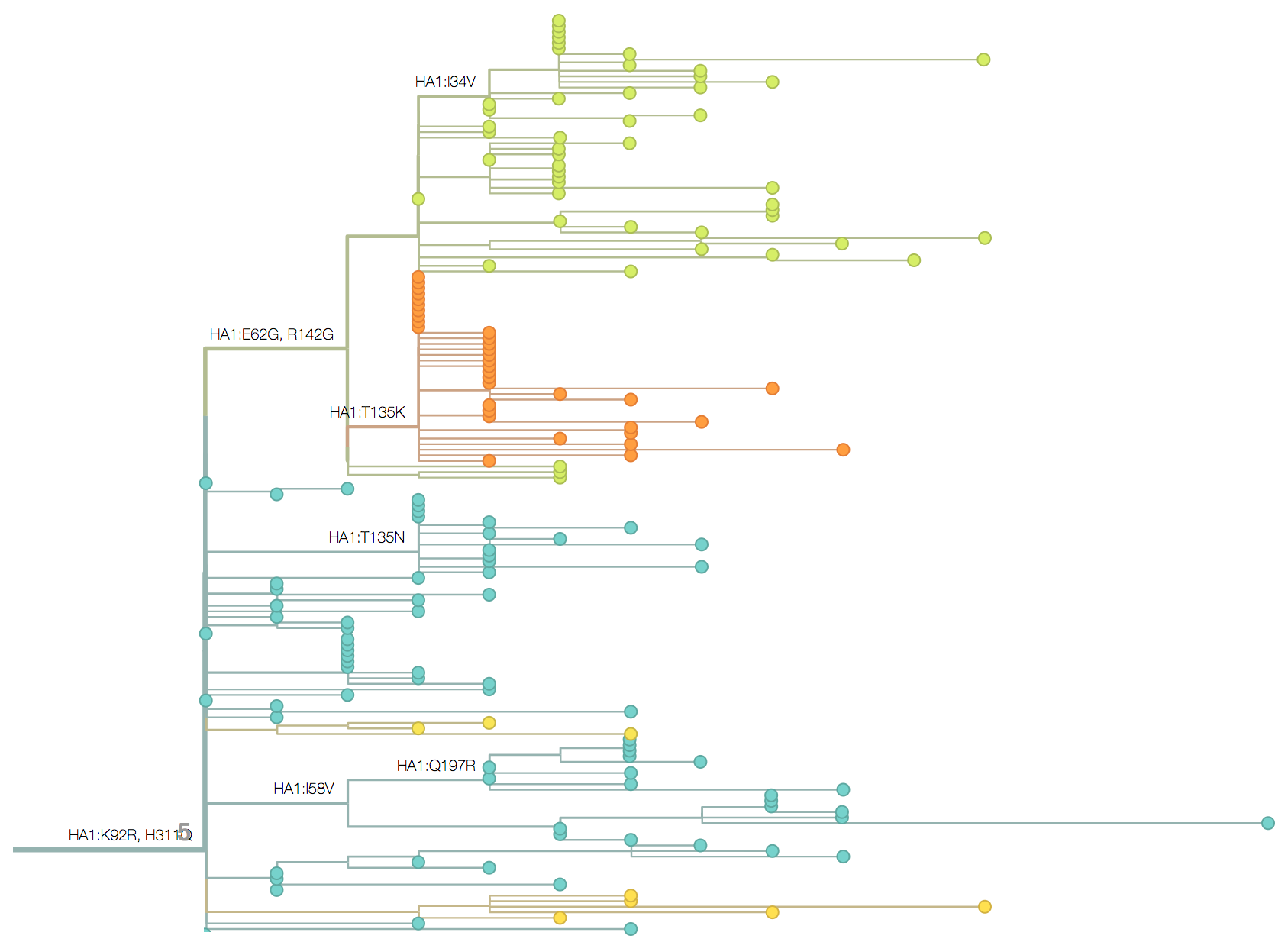
Molecular clocks and dated phylogenies
Mutations tend to accumulate in a clock-like fashion
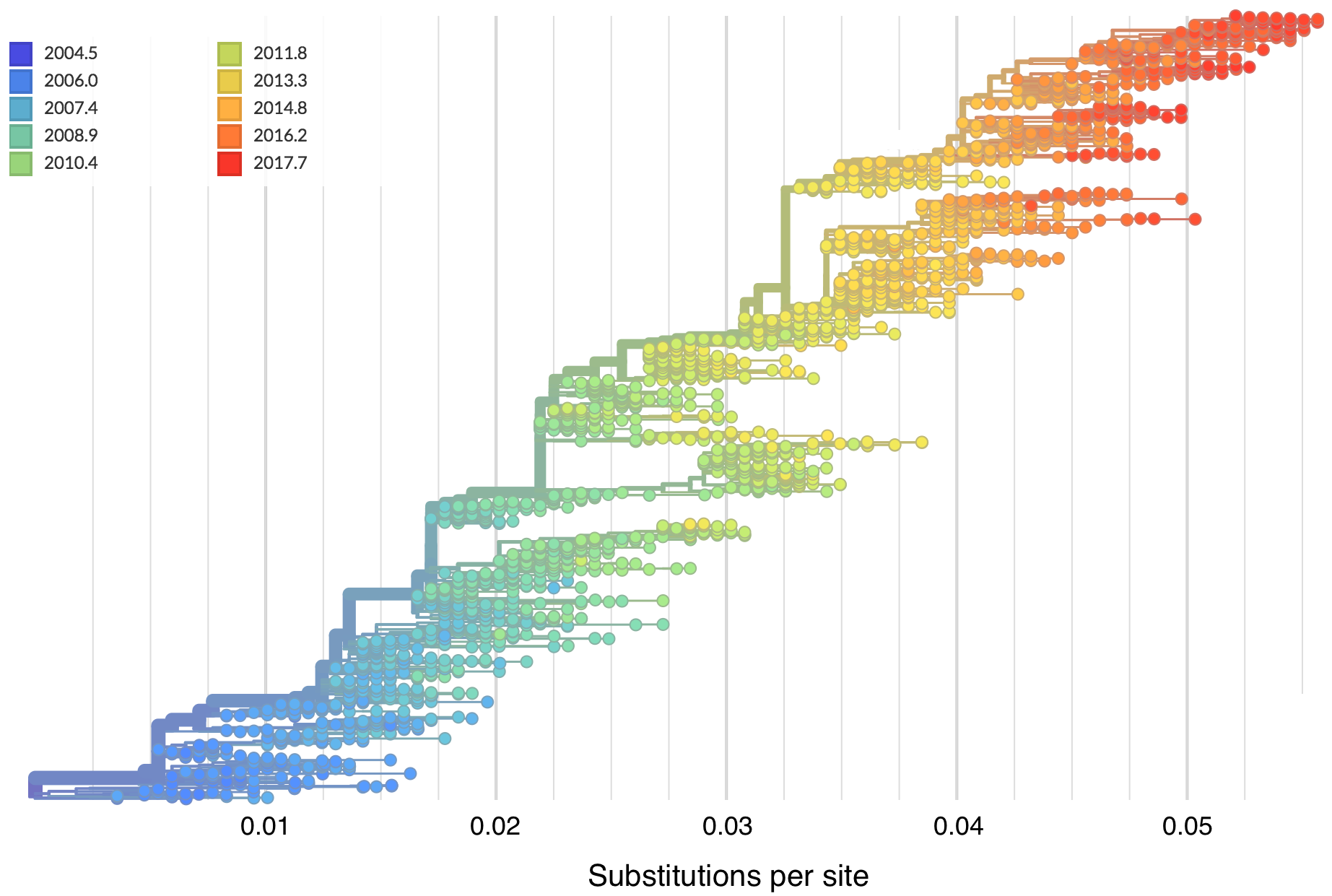
"Root-to-tip" plots show temporal signal
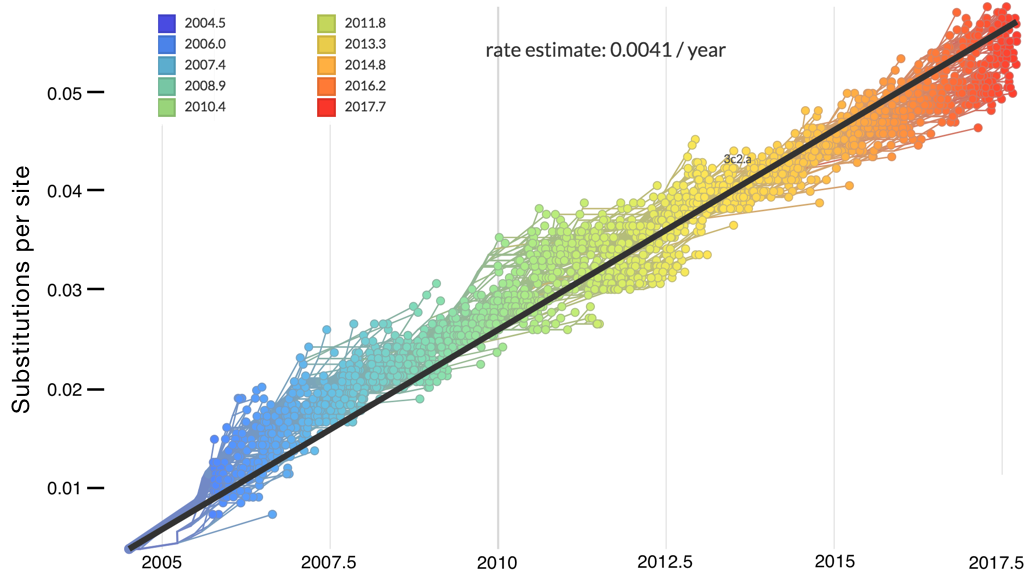
Allows conversion between branch length and time
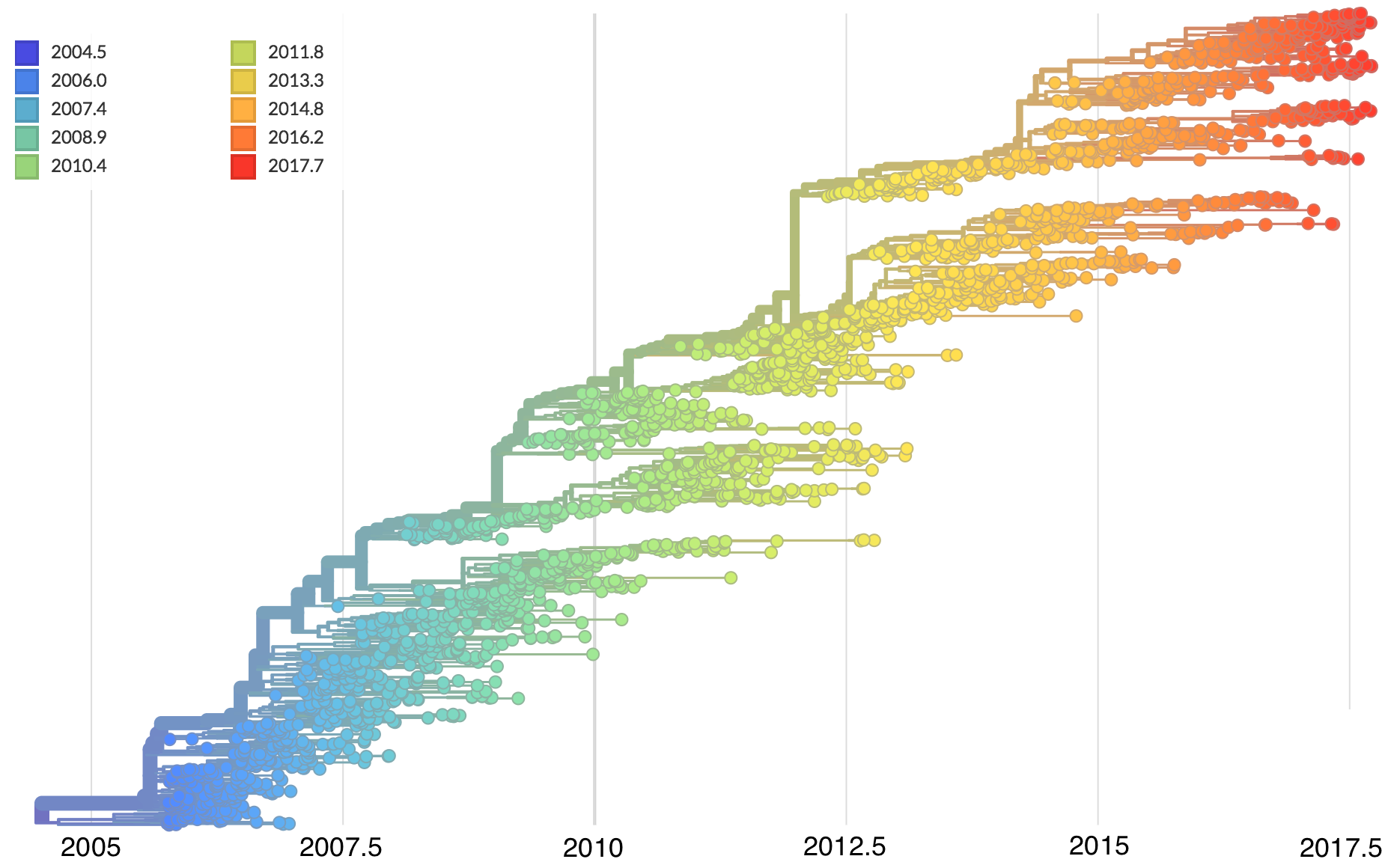
Dated phylogenies provide real-world context
Inference of discrete traits
"Data" is a phylogeny and tip states
States include nucleotides, amino acids, geo locations, hosts, etc...
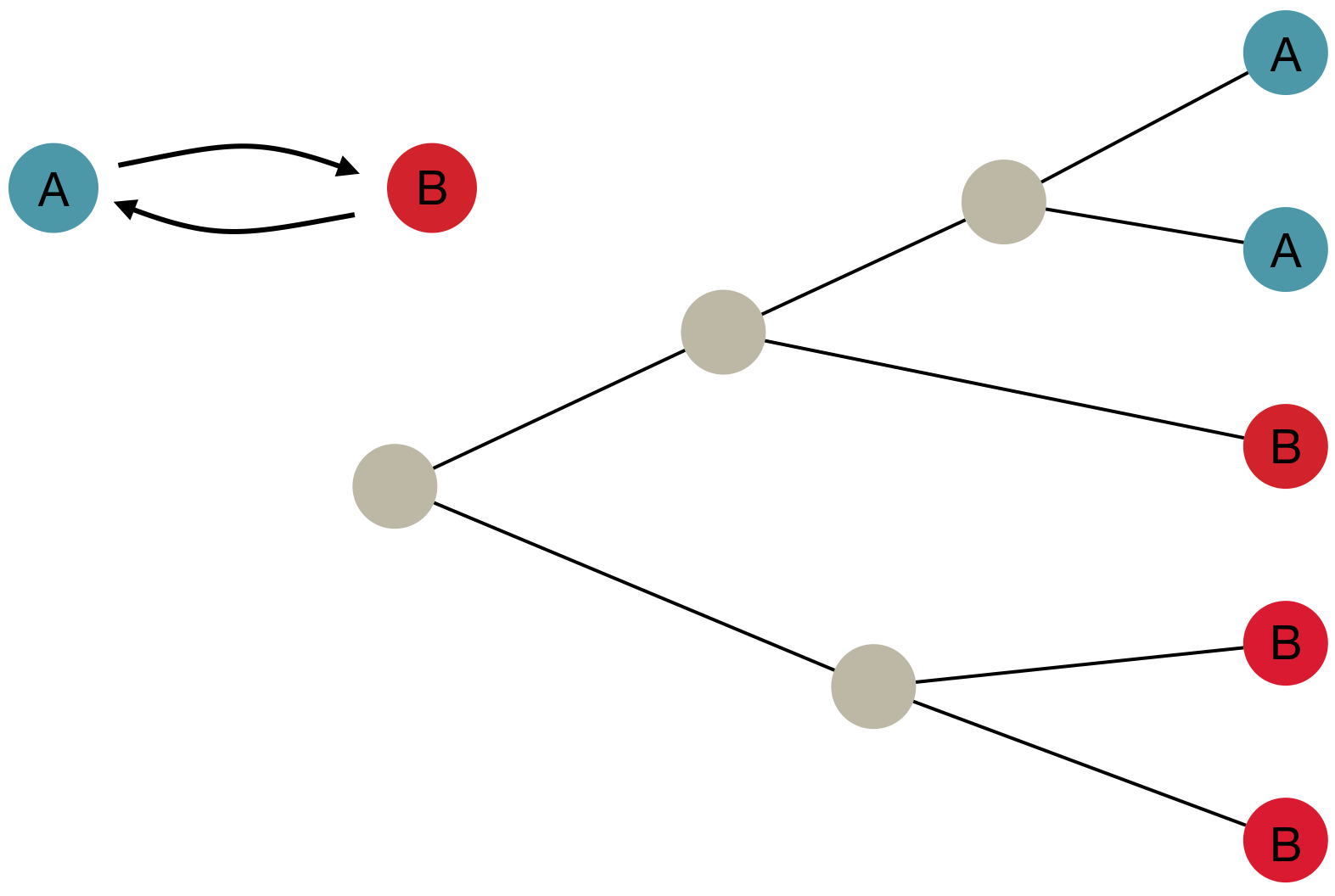
Model infers transition matrix and ancestral states

Rare transitions, short branches and many taxa increase confidence

Phylogeography
Nesting patterns are informative
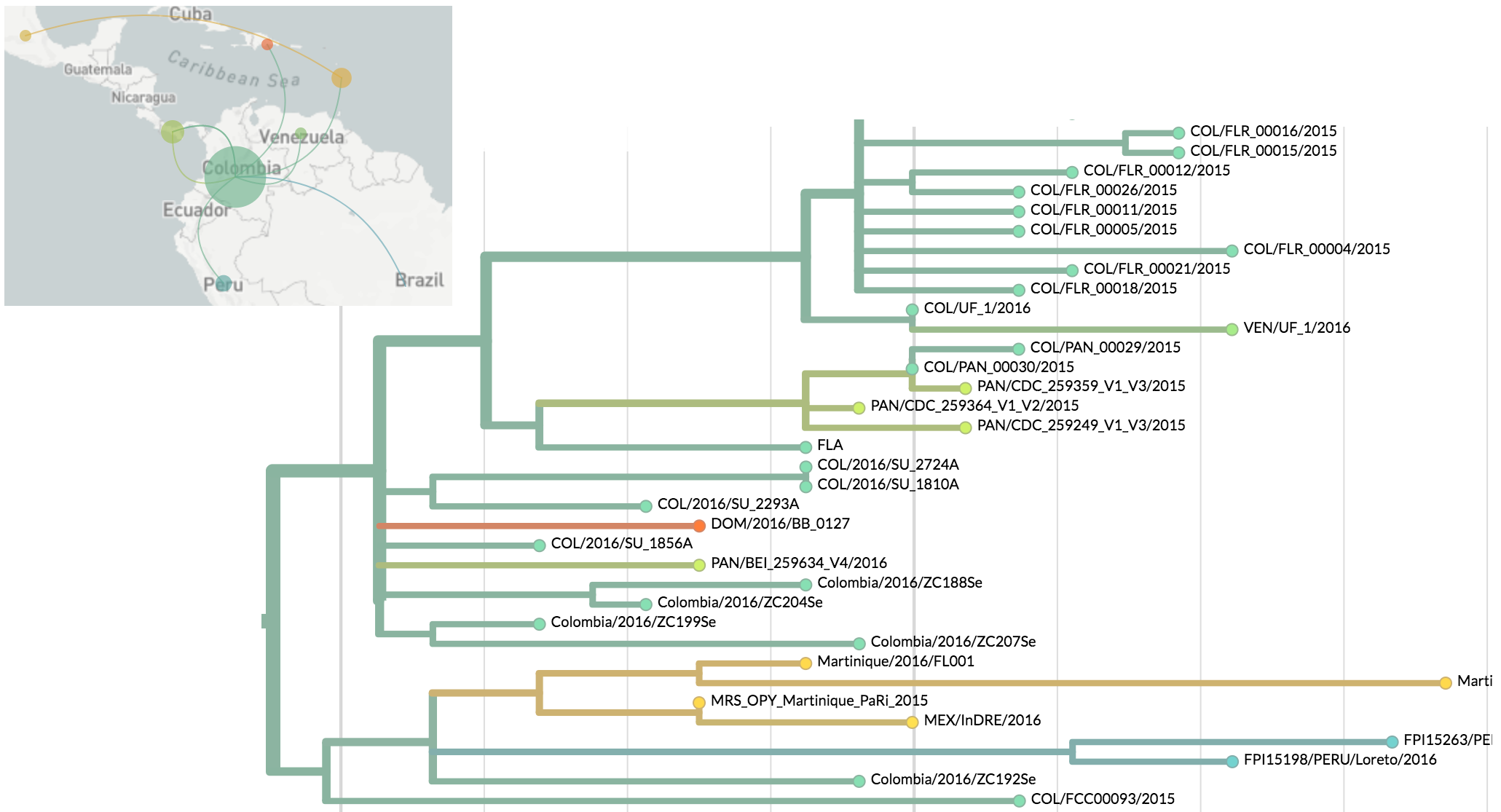
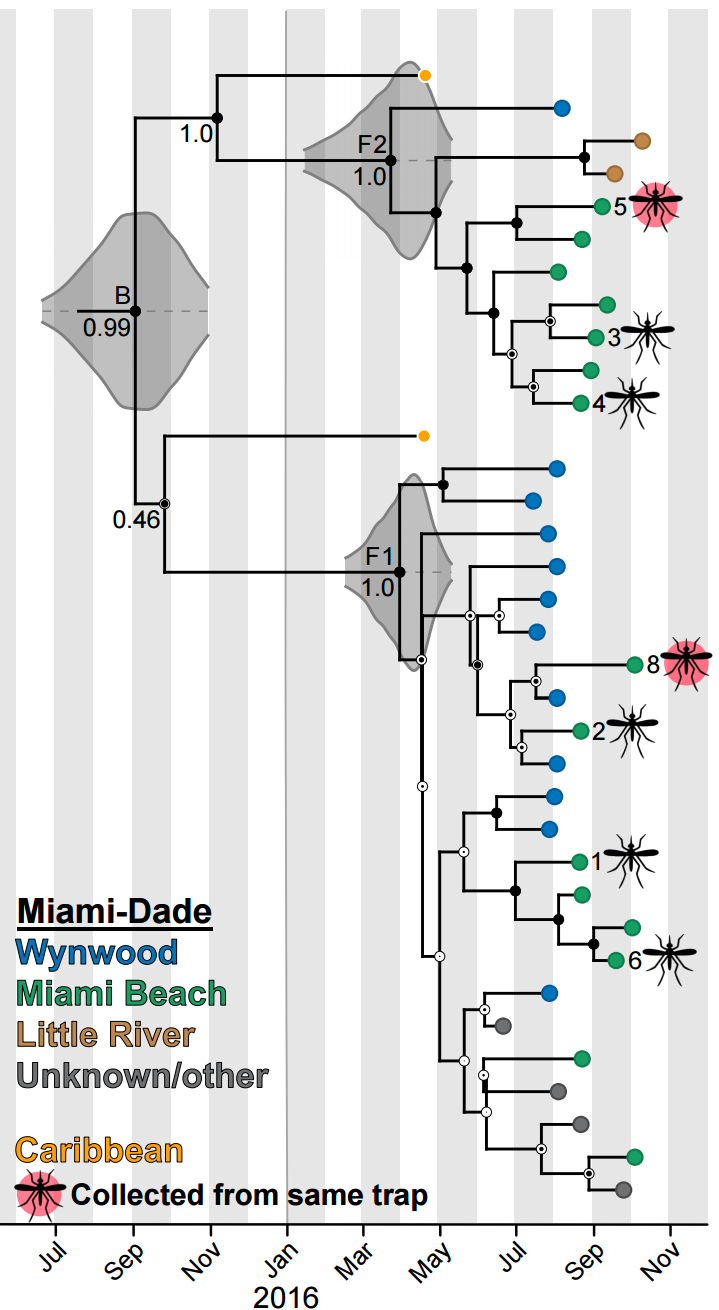
Zika phylogeny infers an origin in northeast Brazil
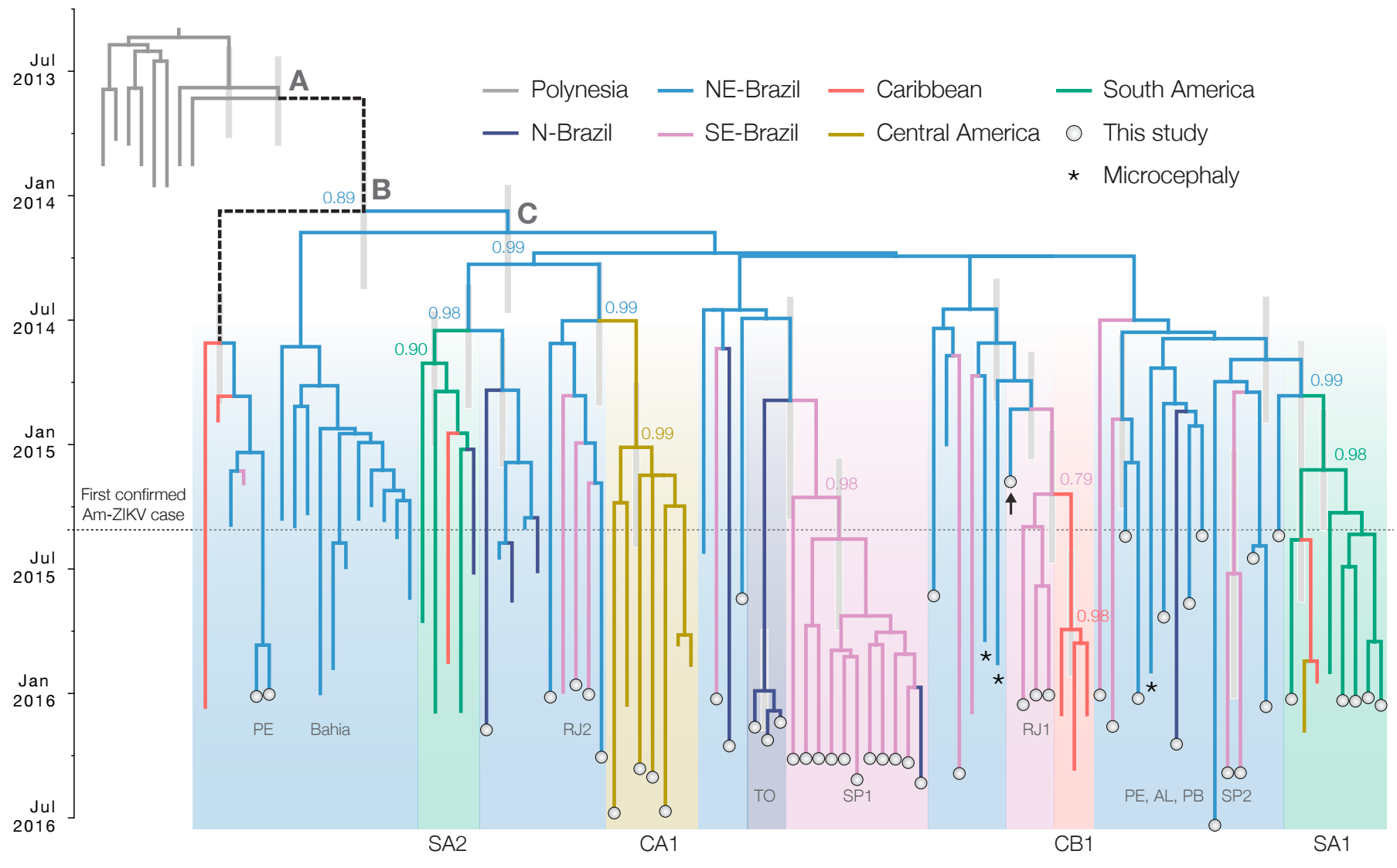
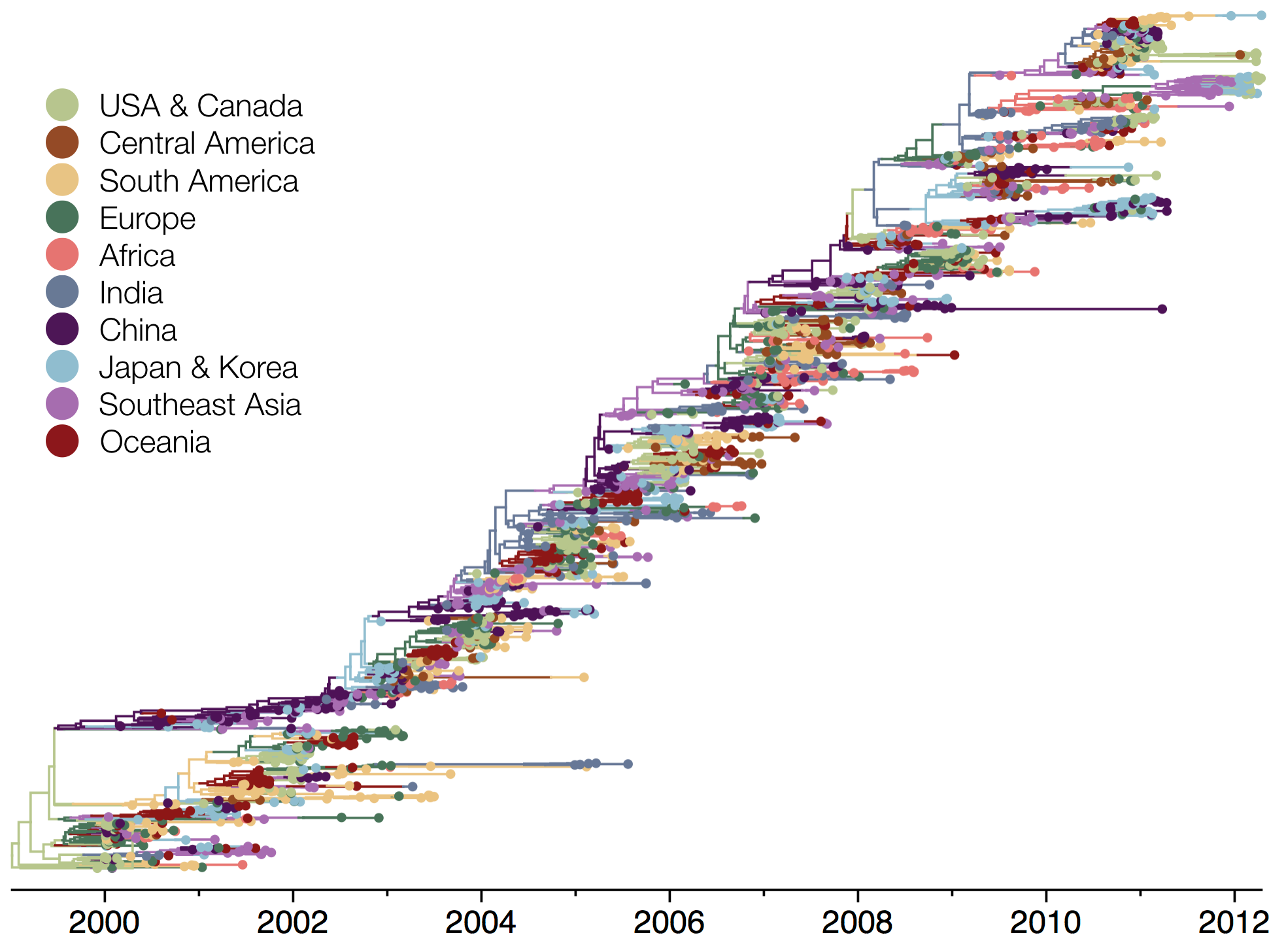
Influenza phylogeny shows repeated spread from E-SE Asia
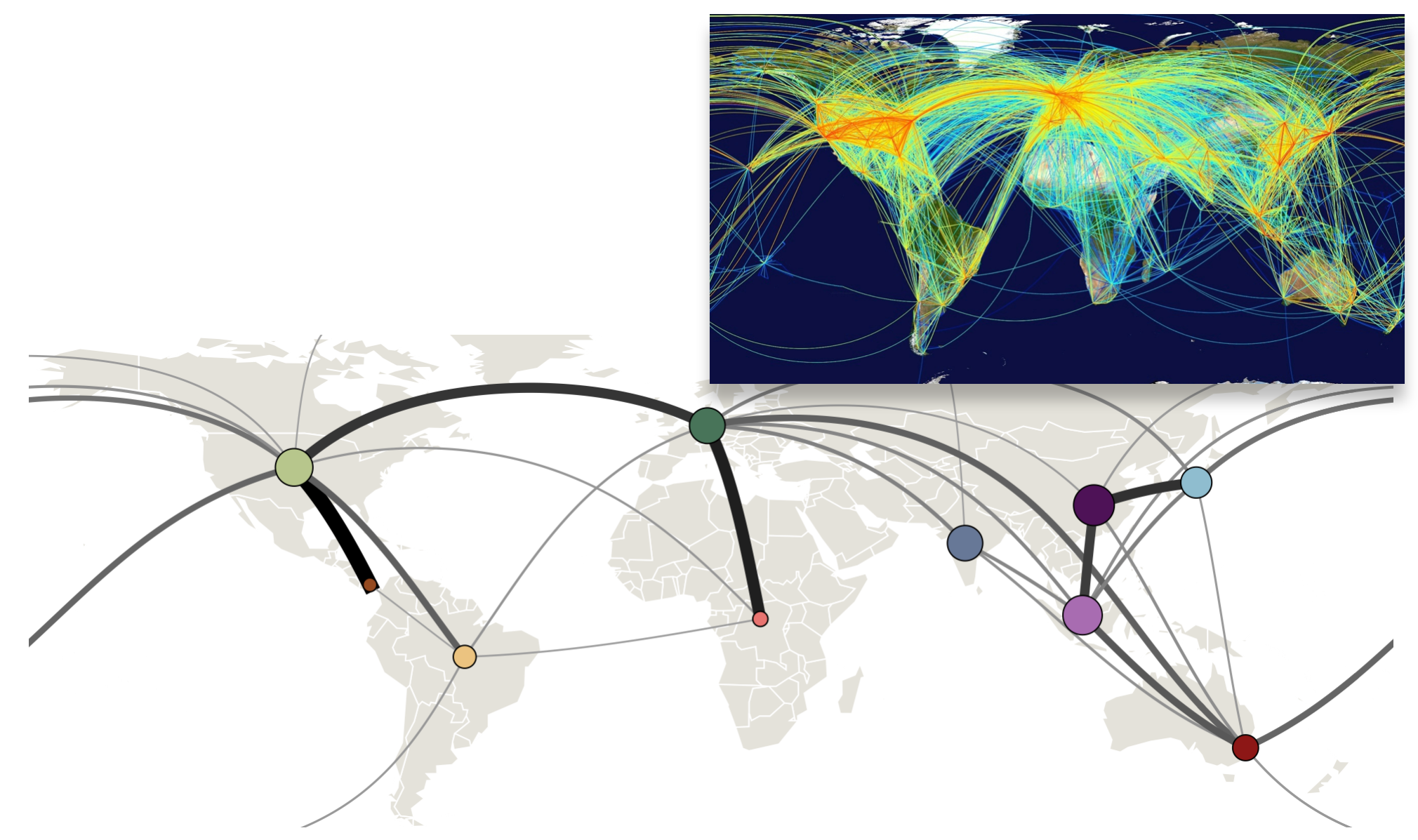
Influenza transitions mirror air travel network
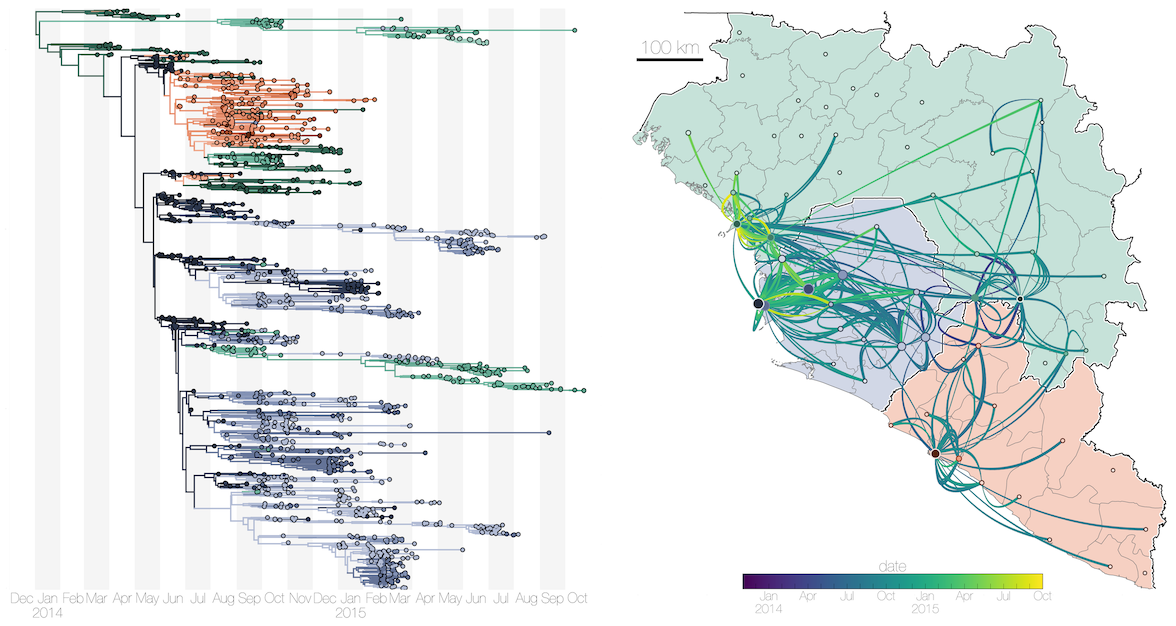
Ebola phylogeny shows frequent migration events
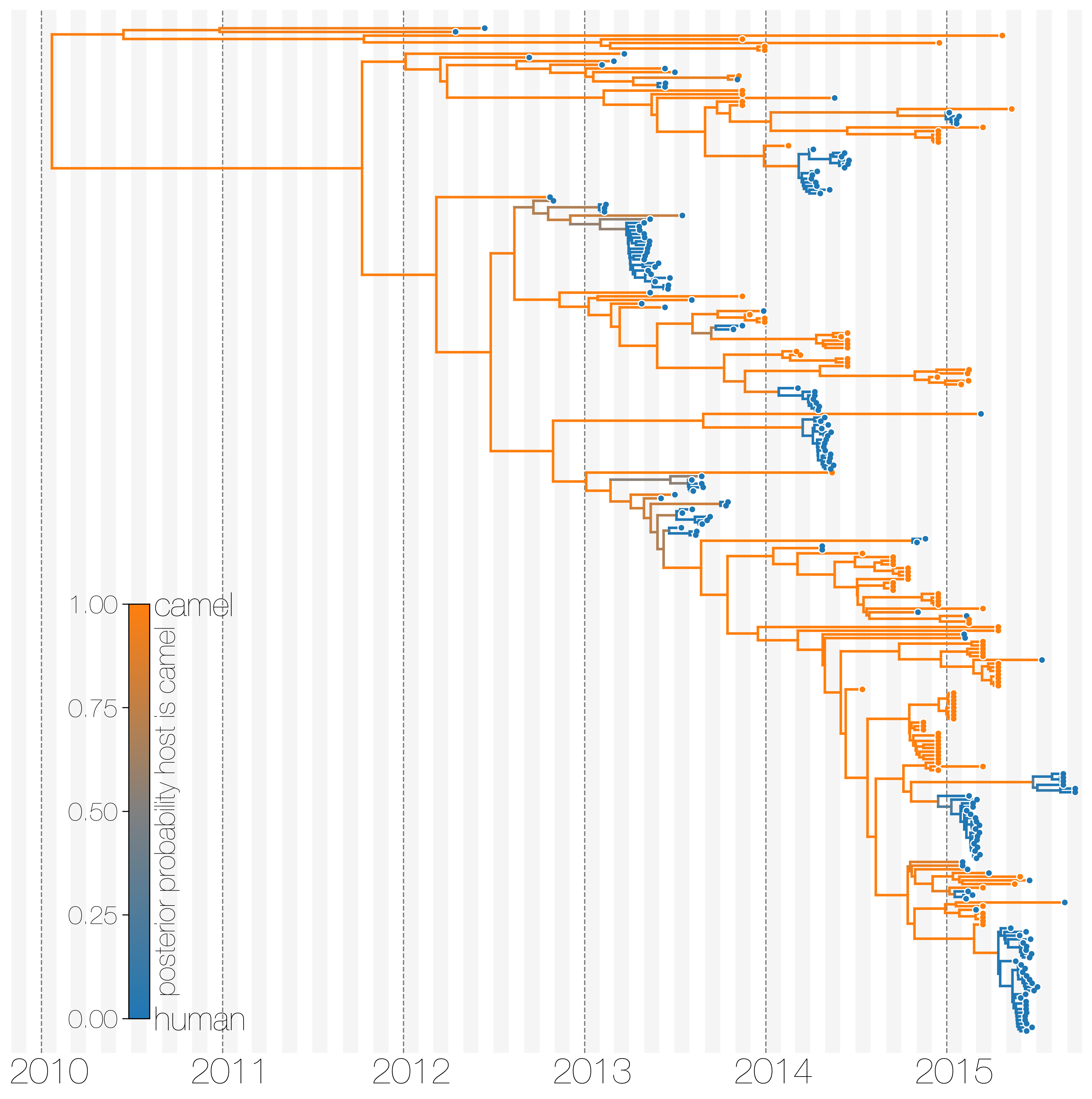
Reservoir species and host jumps
MERS-CoV has frequent spillover events, but limited human-to-human transmission
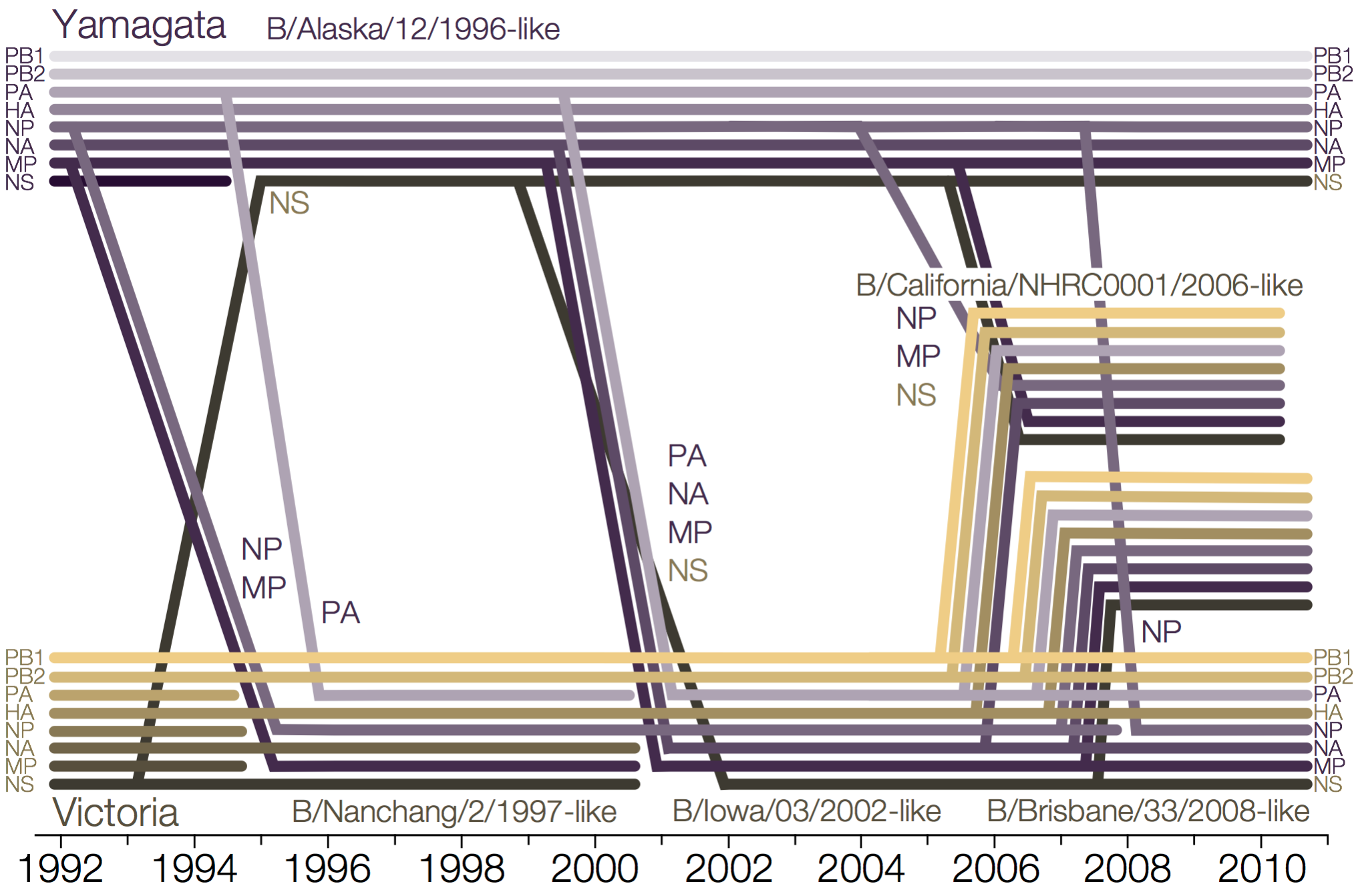
Reassortment and recombination
Influenza B reassorts across segments
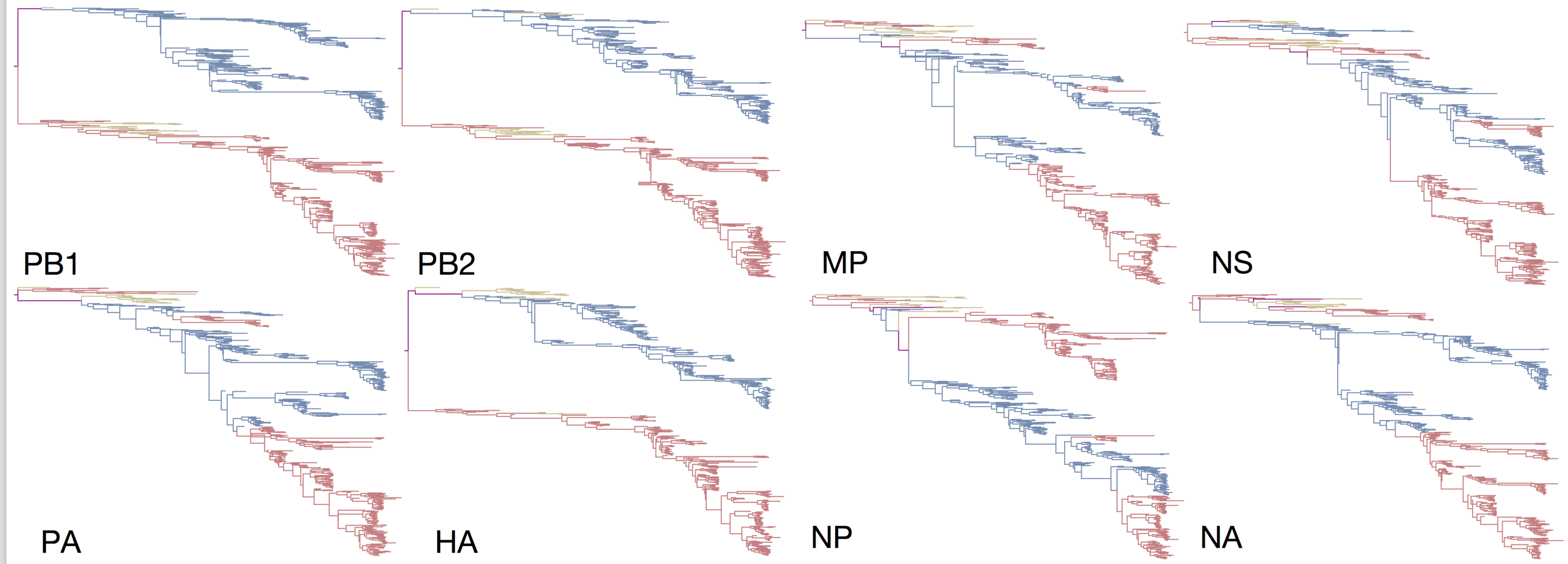
Reassortment splits gene constellations
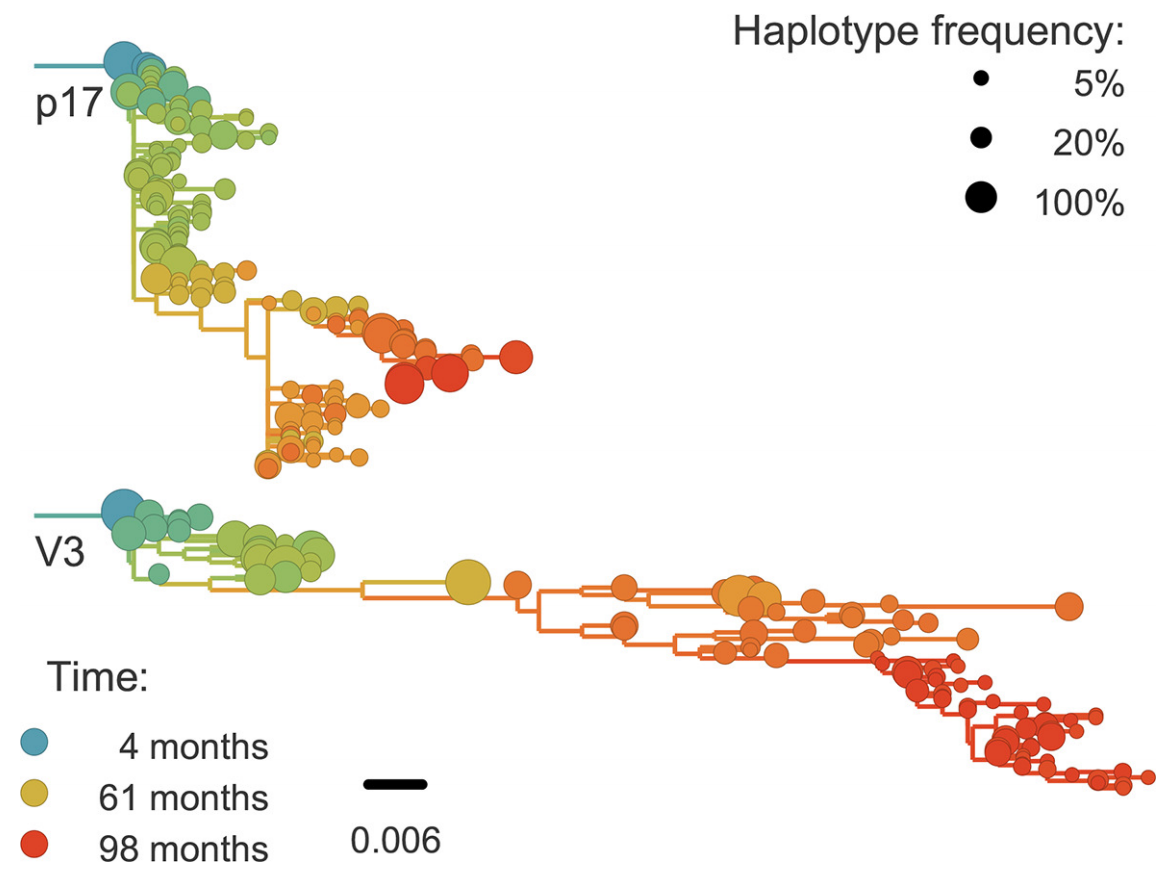
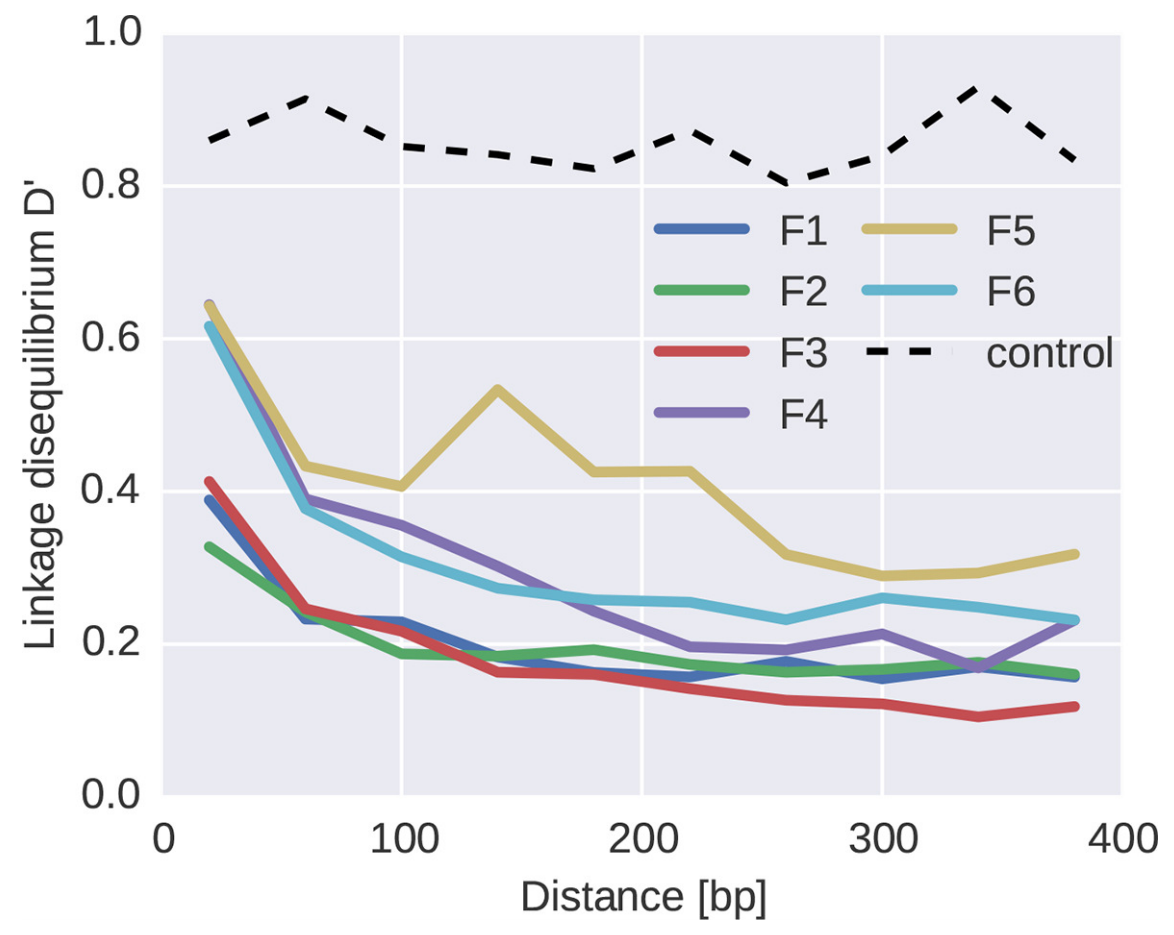
Abundant recombination during within-host HIV evolution
Linkage disequilibrium decays with physical distance
Summary
- Phylogenetics uses sequence data to infer trees along with ancestral states
- Molecular clocks provide a real-world context to evolutionary events
- Phylogeny reveals behavior of discrete traits, like geographic location and host state
- Reassortment / recombination results in non-tree-like evolutionary relationships