Discrete traits and phylogeography
Trevor Bedford (@trvrb)
May 28, 2020
GS541 Introduction to Computational Molecular Biology
Recap
Mutations tend to accumulate in a clock-like fashion
Allows conversion between branch length and time
Population dynamics inferred through coalescence through time
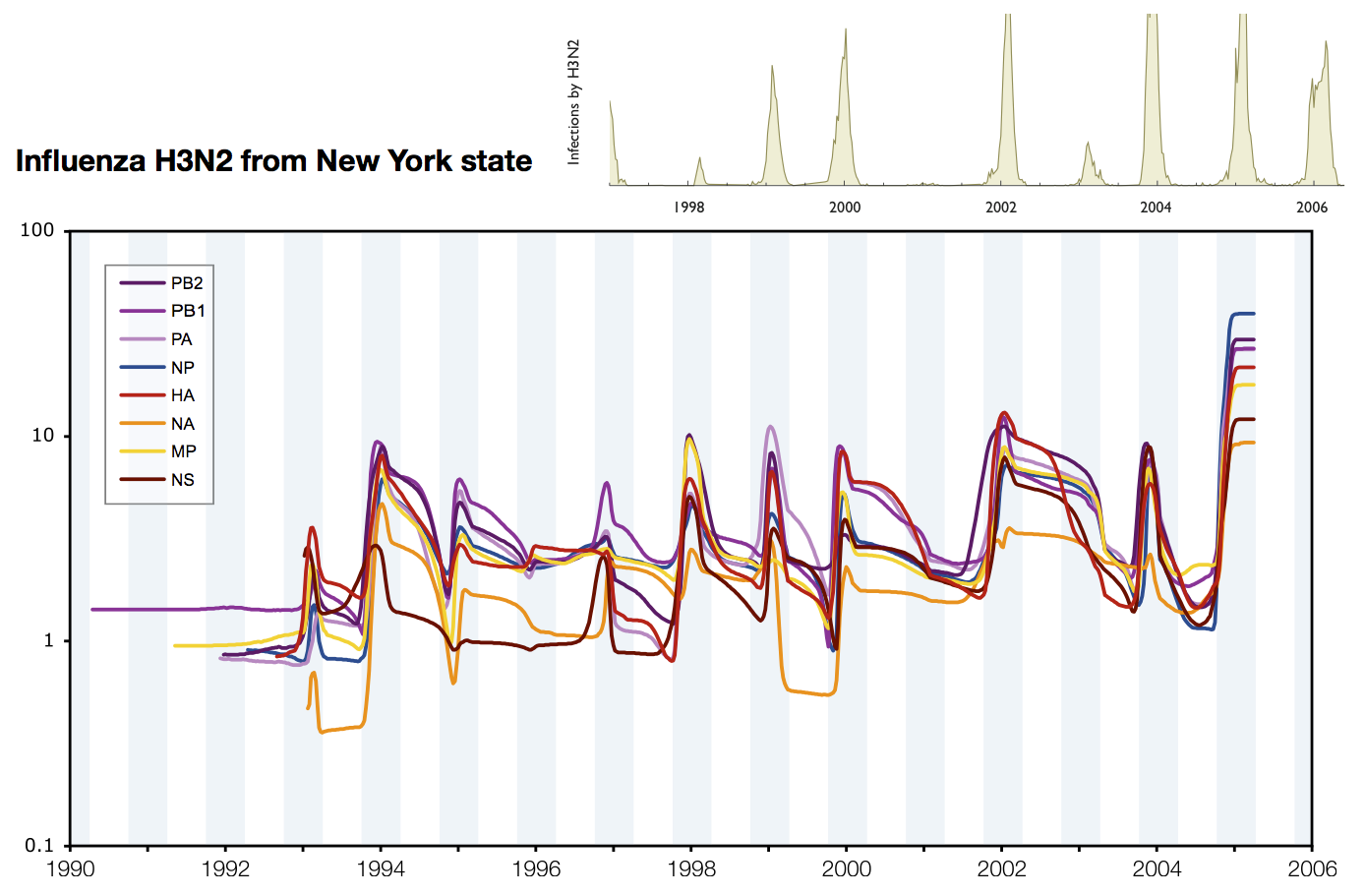
Skyline model shows seasonality in flu
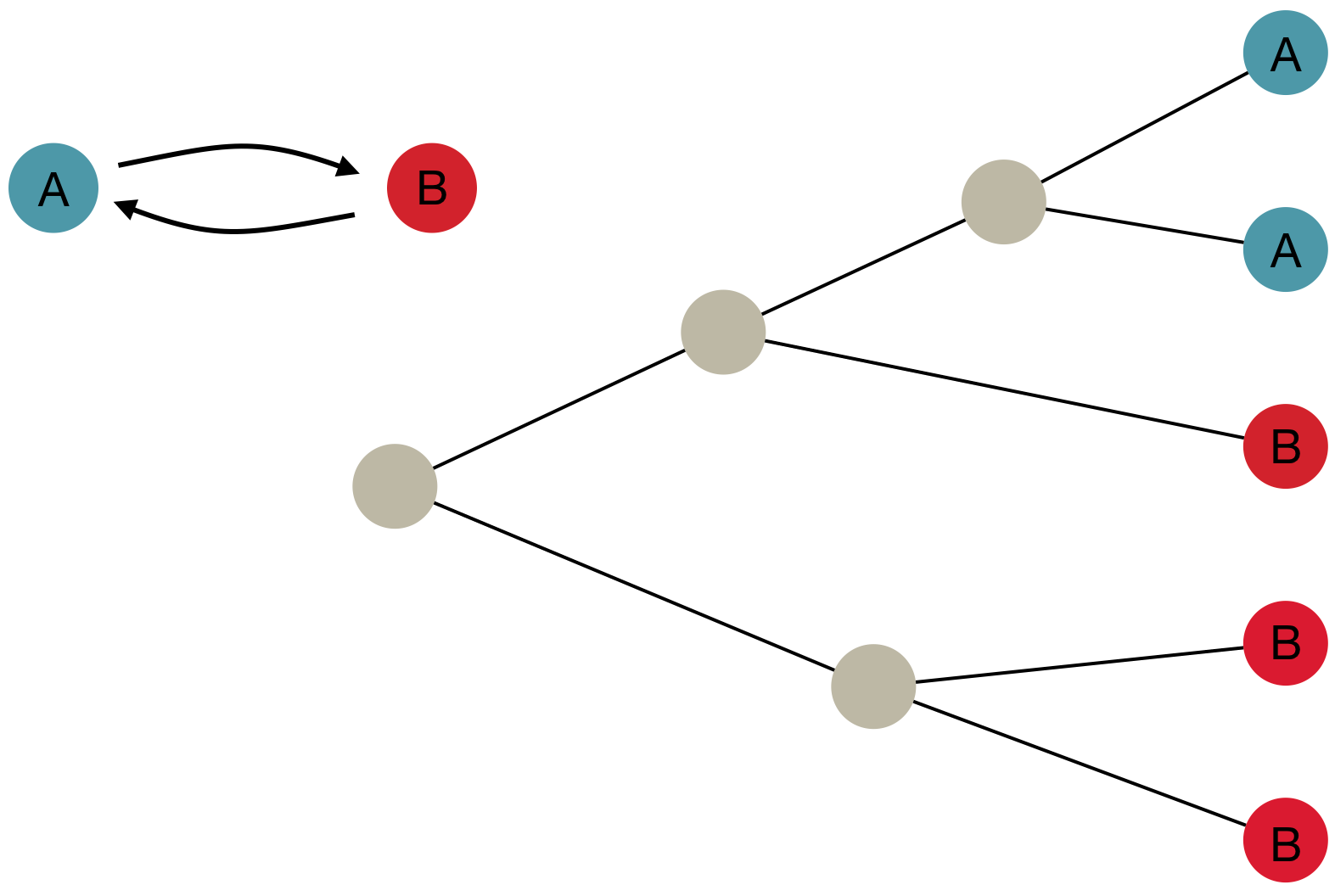
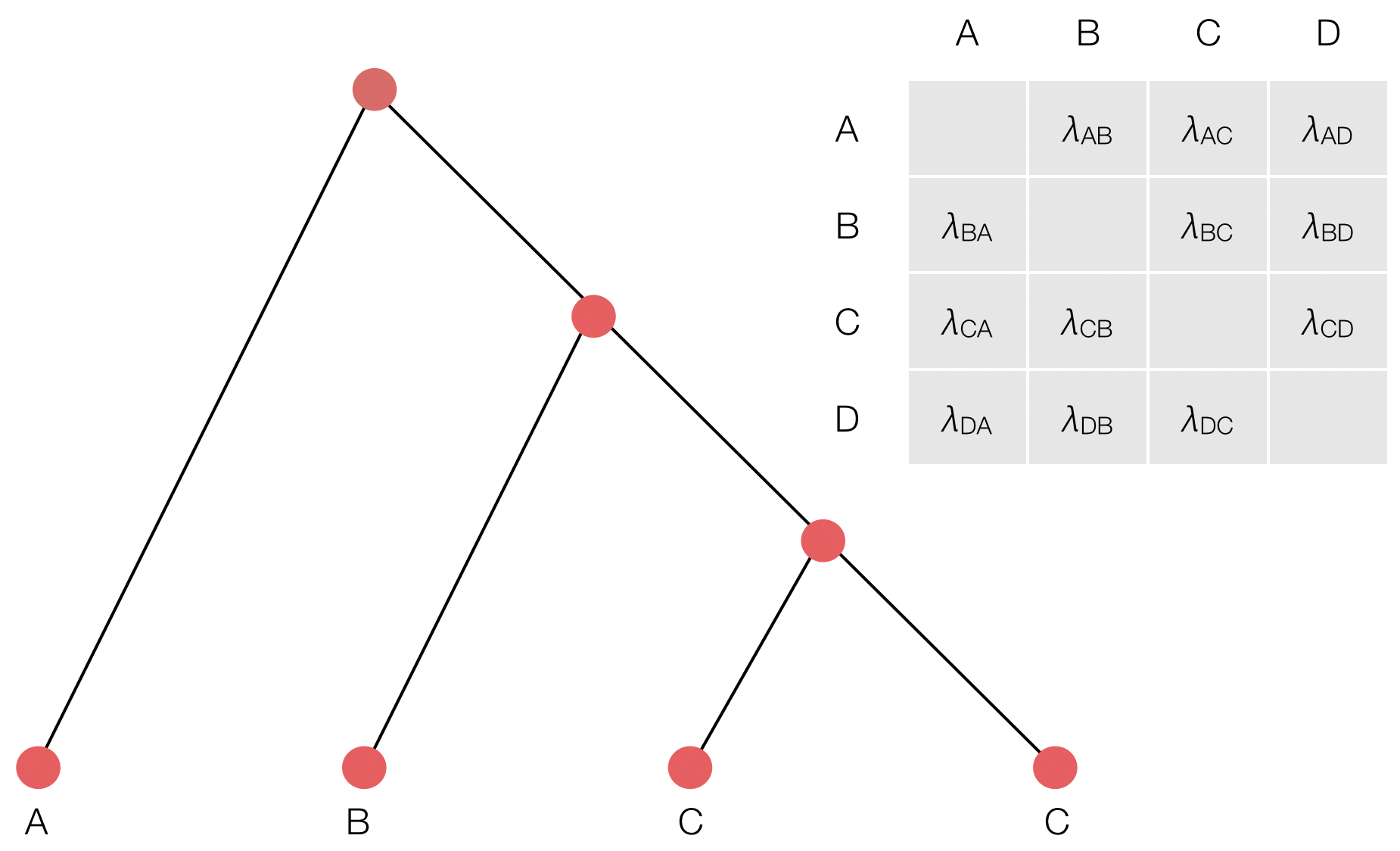
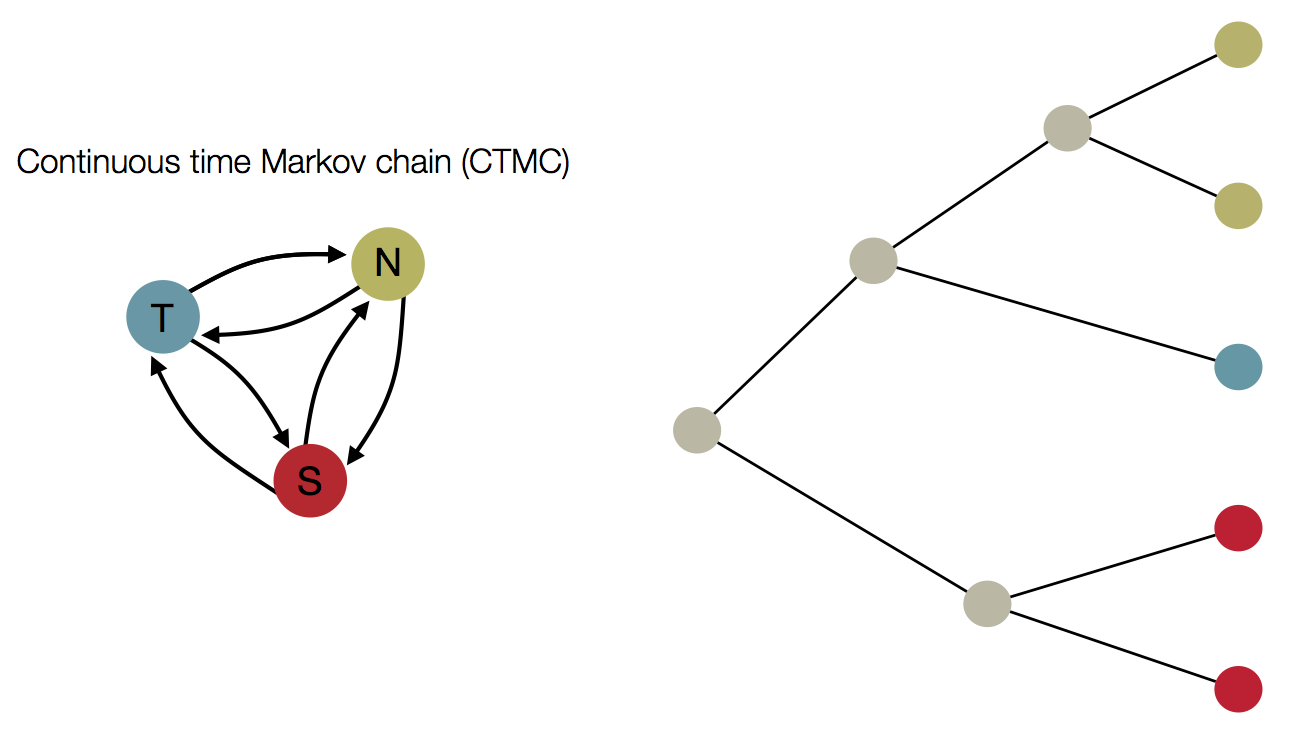
Inference of discrete traits
"Data" is a phylogeny and tip states
States include nucleotides, amino acids, geo locations, hosts, etc...
Model infers transition matrix and ancestral states
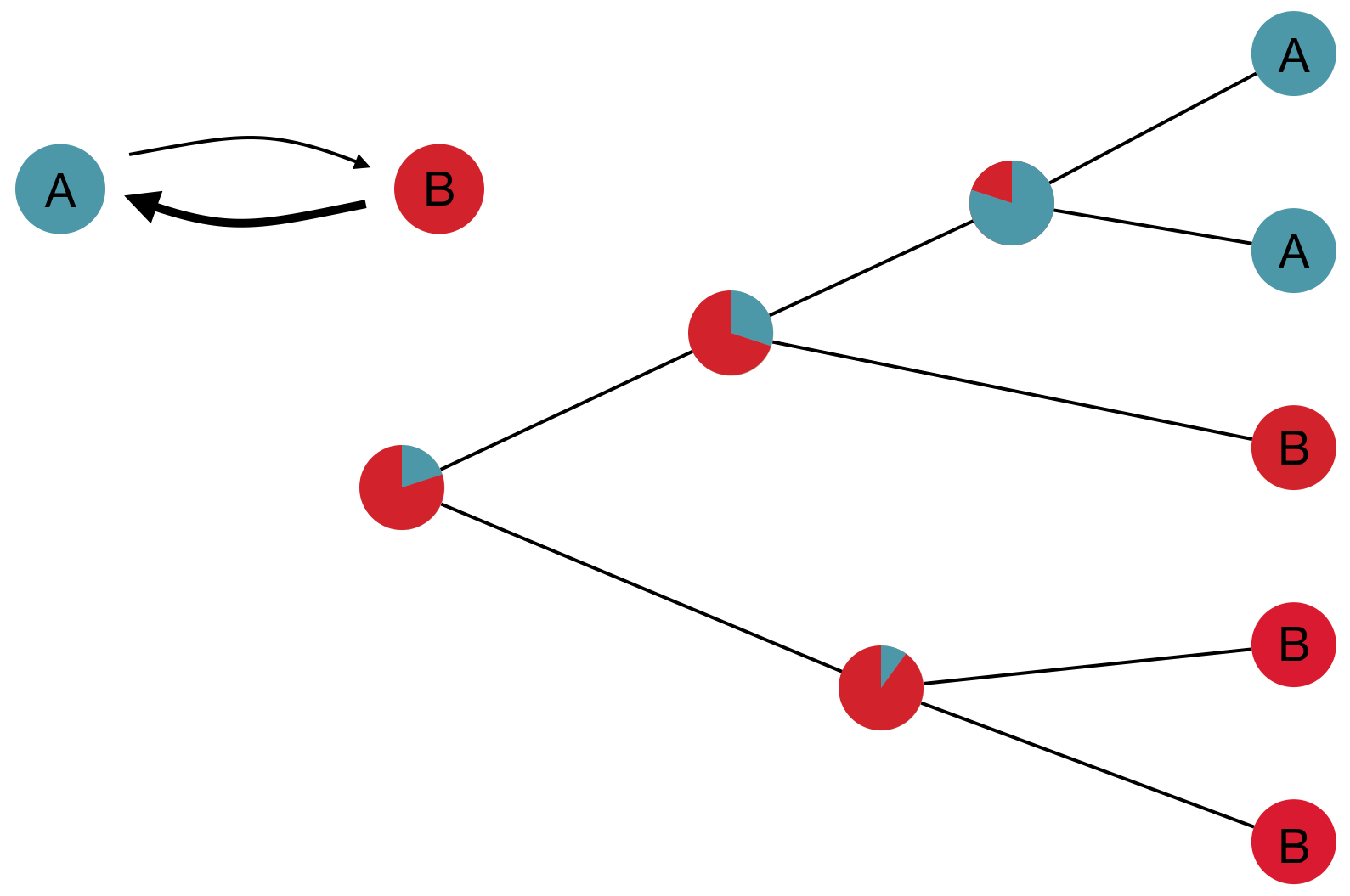
Rare transitions, short branches and many taxa increase confidence
CTMC transition matrix
Likelihood calculations as before
Parameterization of CTMC transition matrix
via Philippe Lemey and Marc Suchard
Phylogeography
Nesting patterns are informative
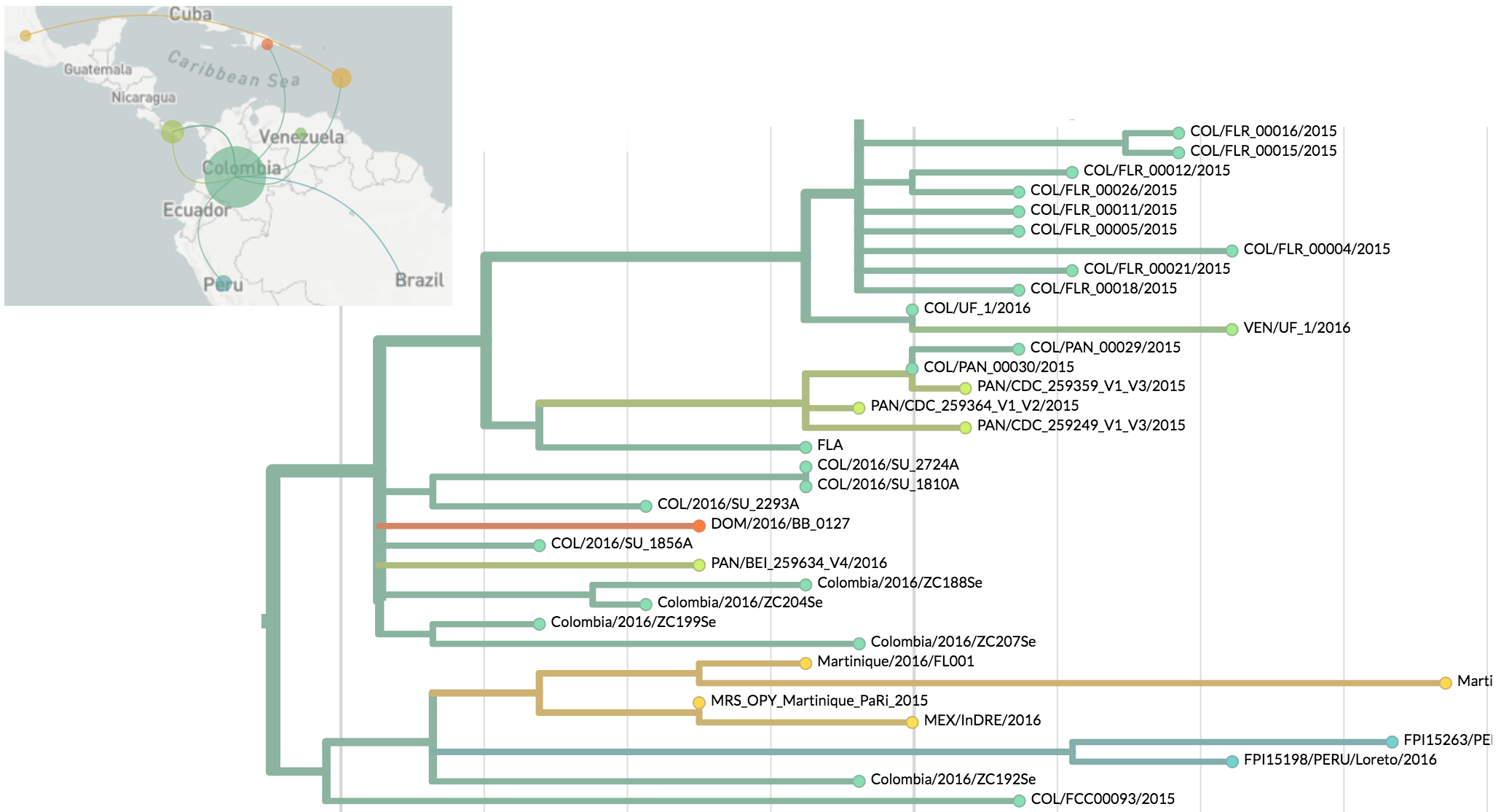
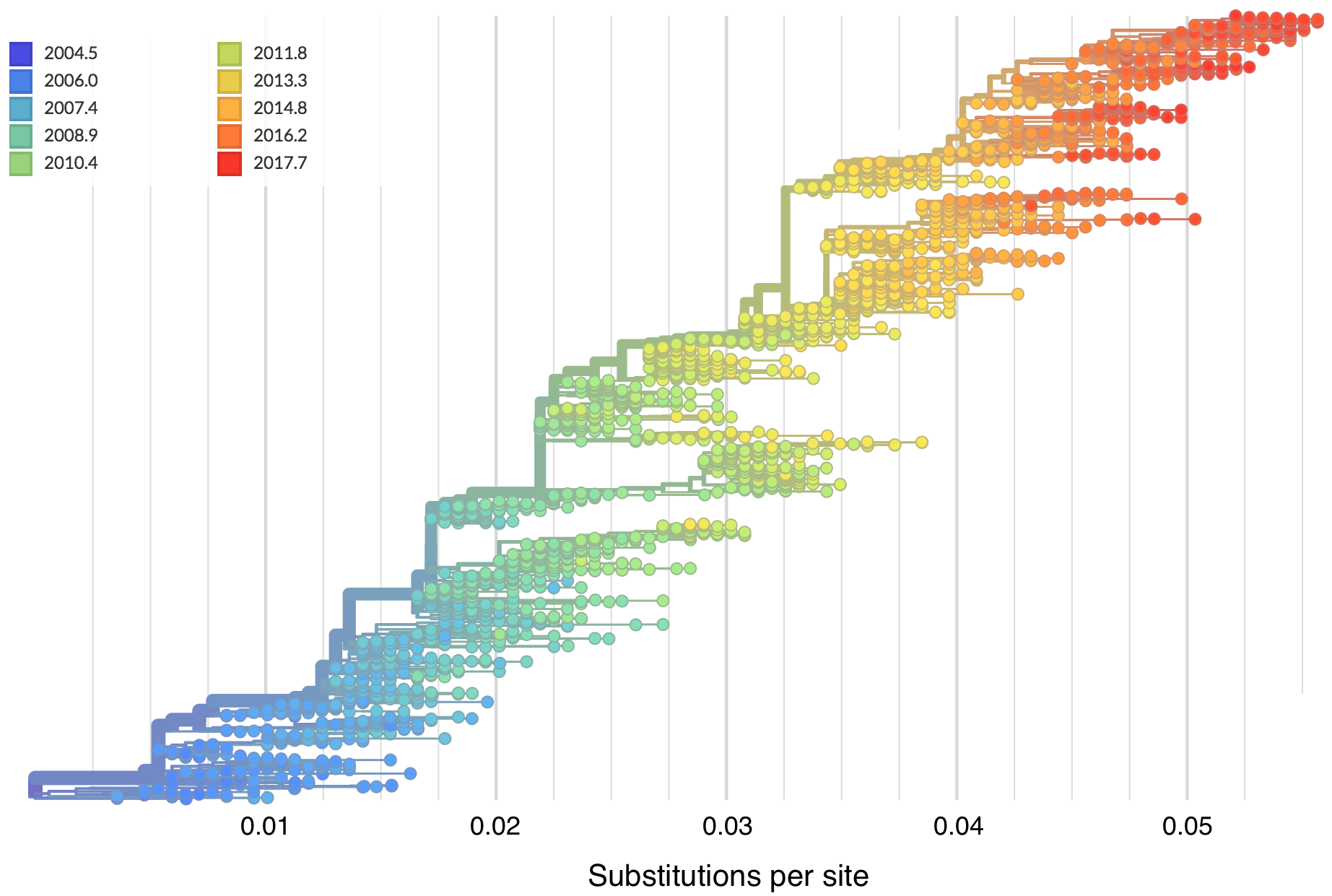
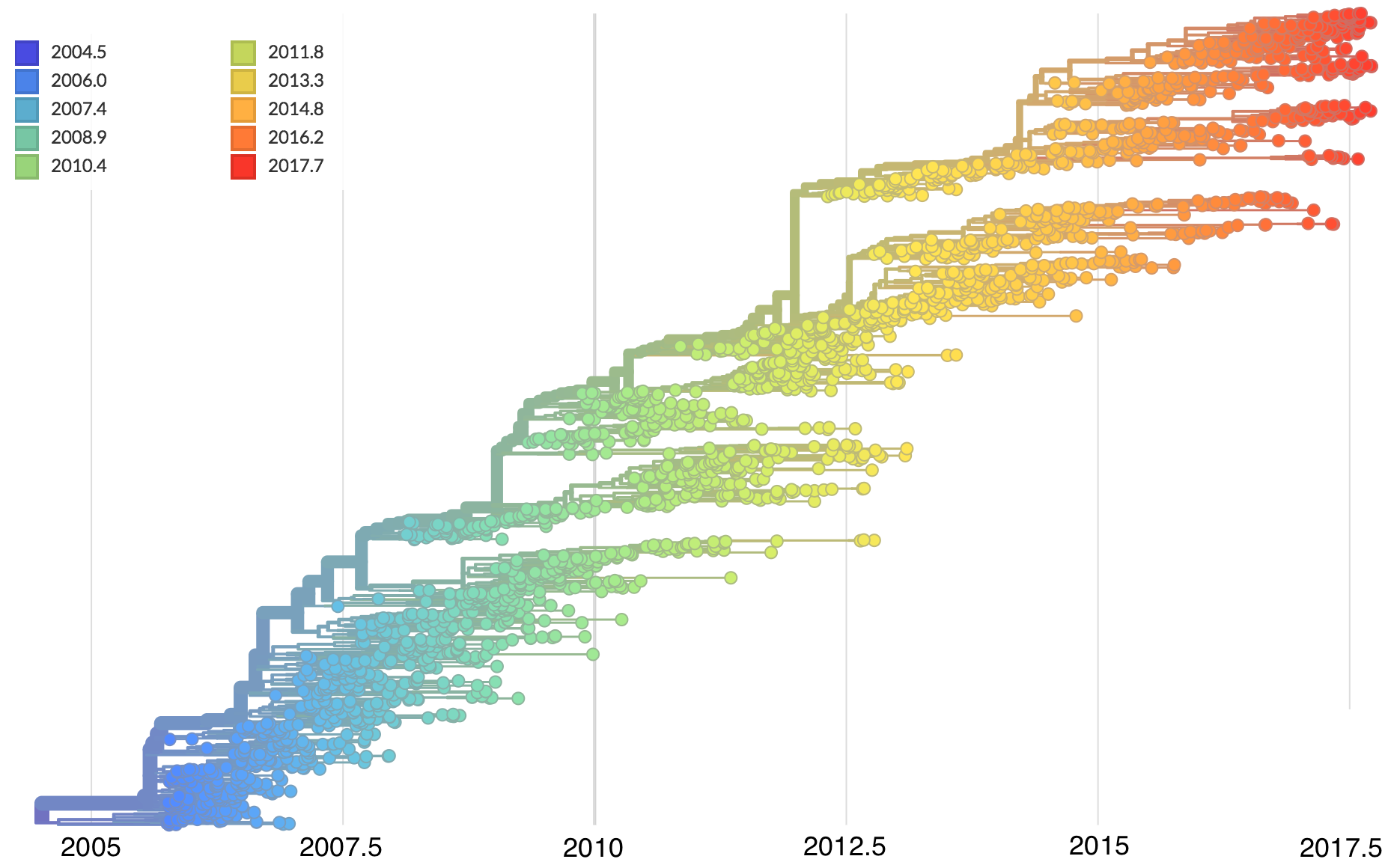
Zika phylogeny infers an origin in northeast Brazil
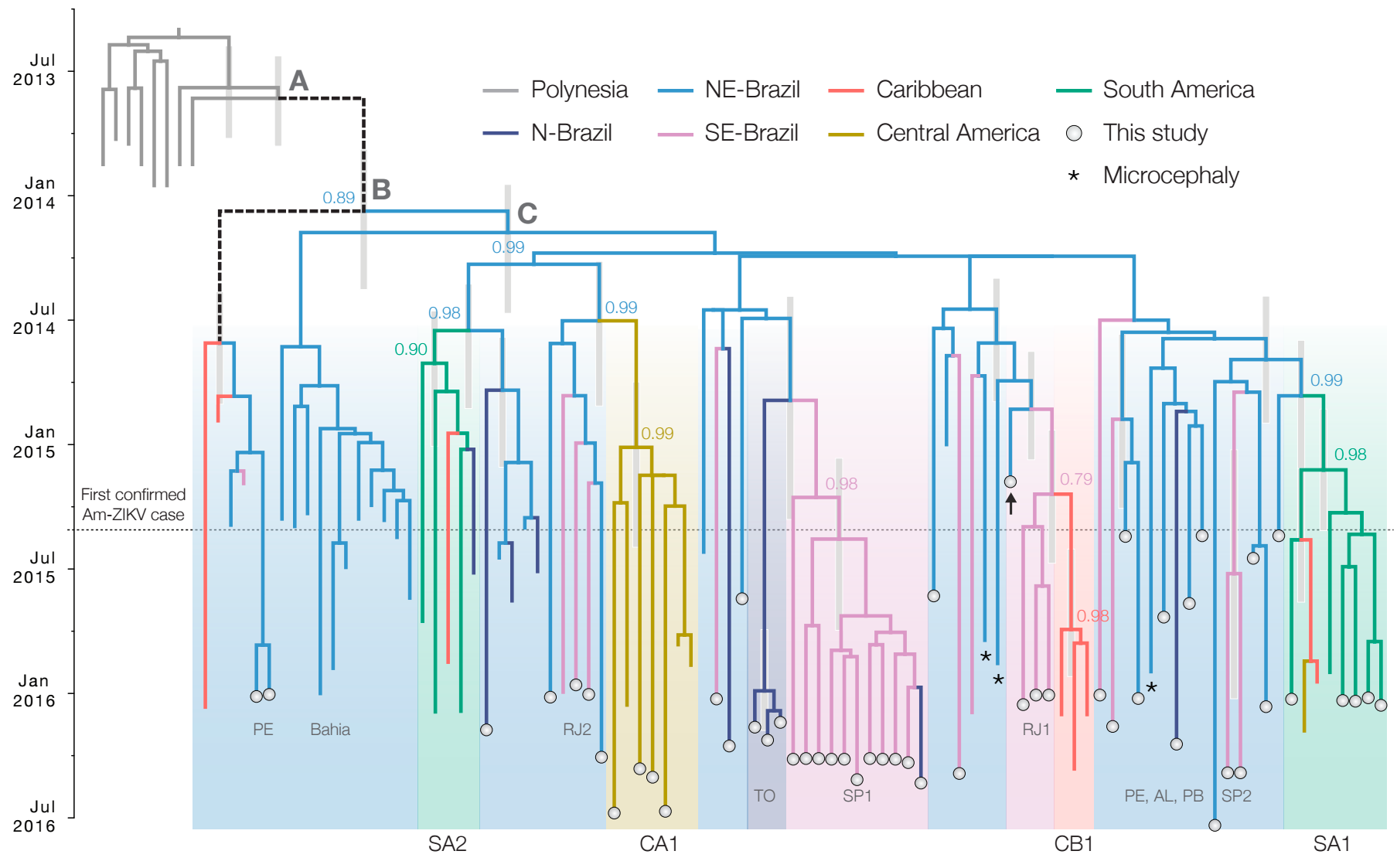
Seasonality in influenza
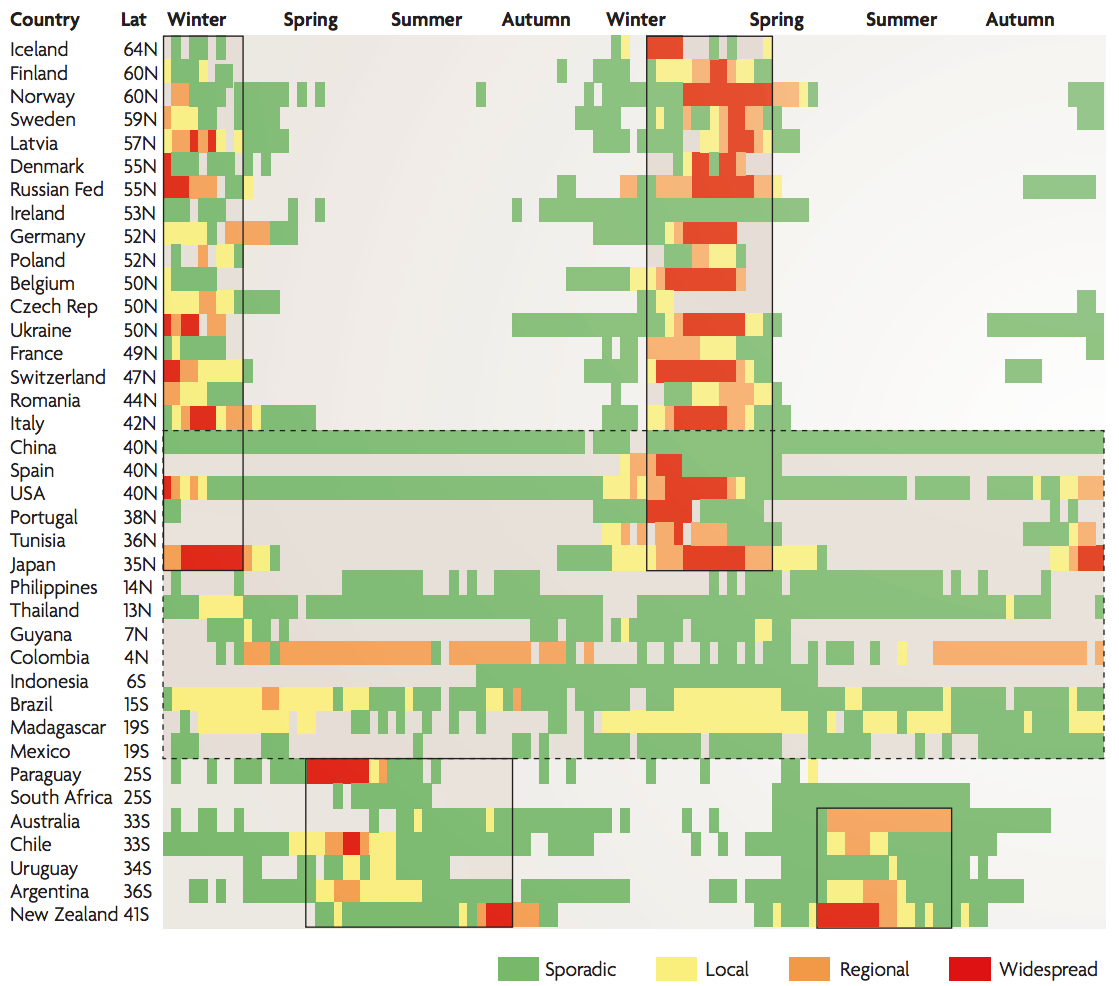
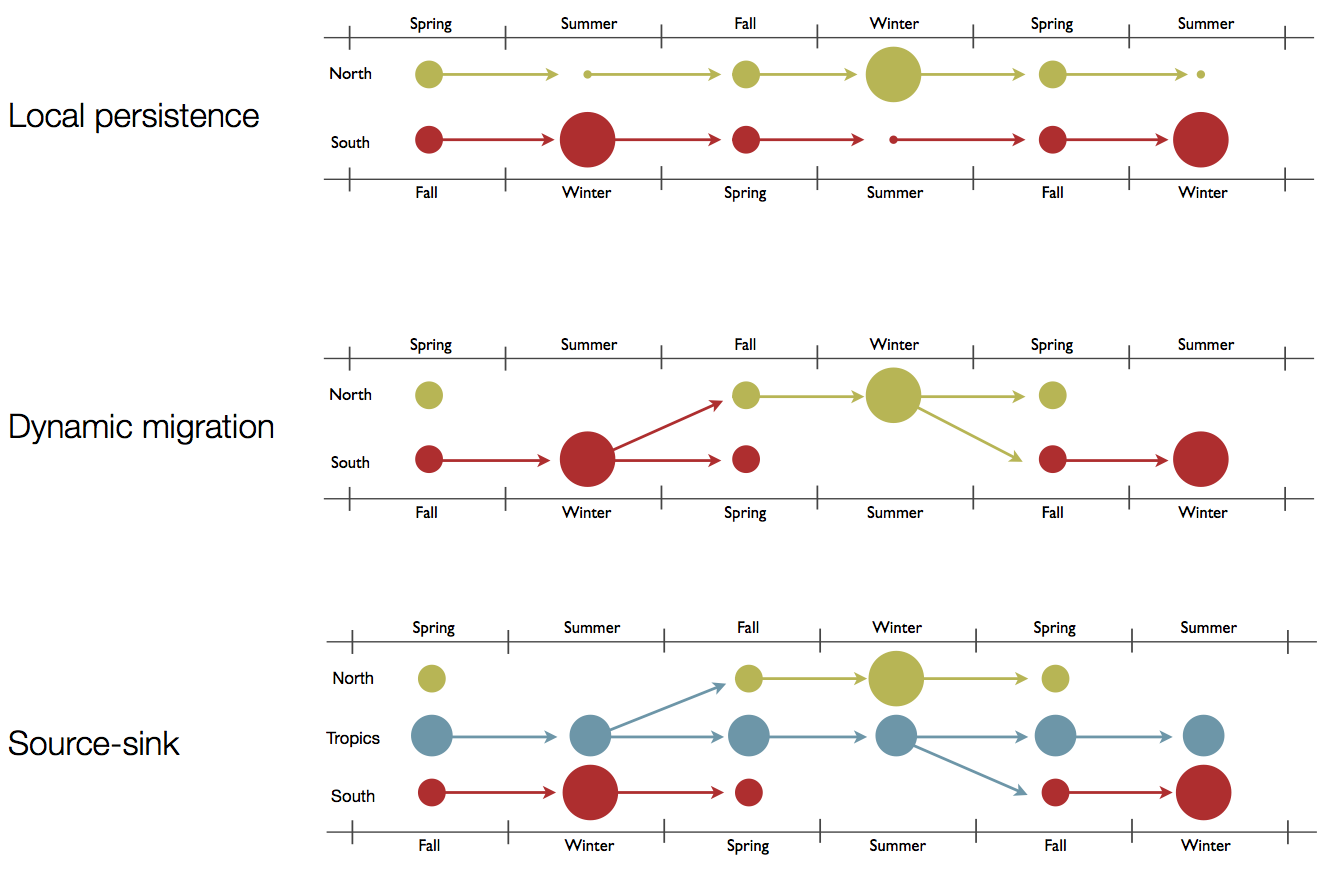
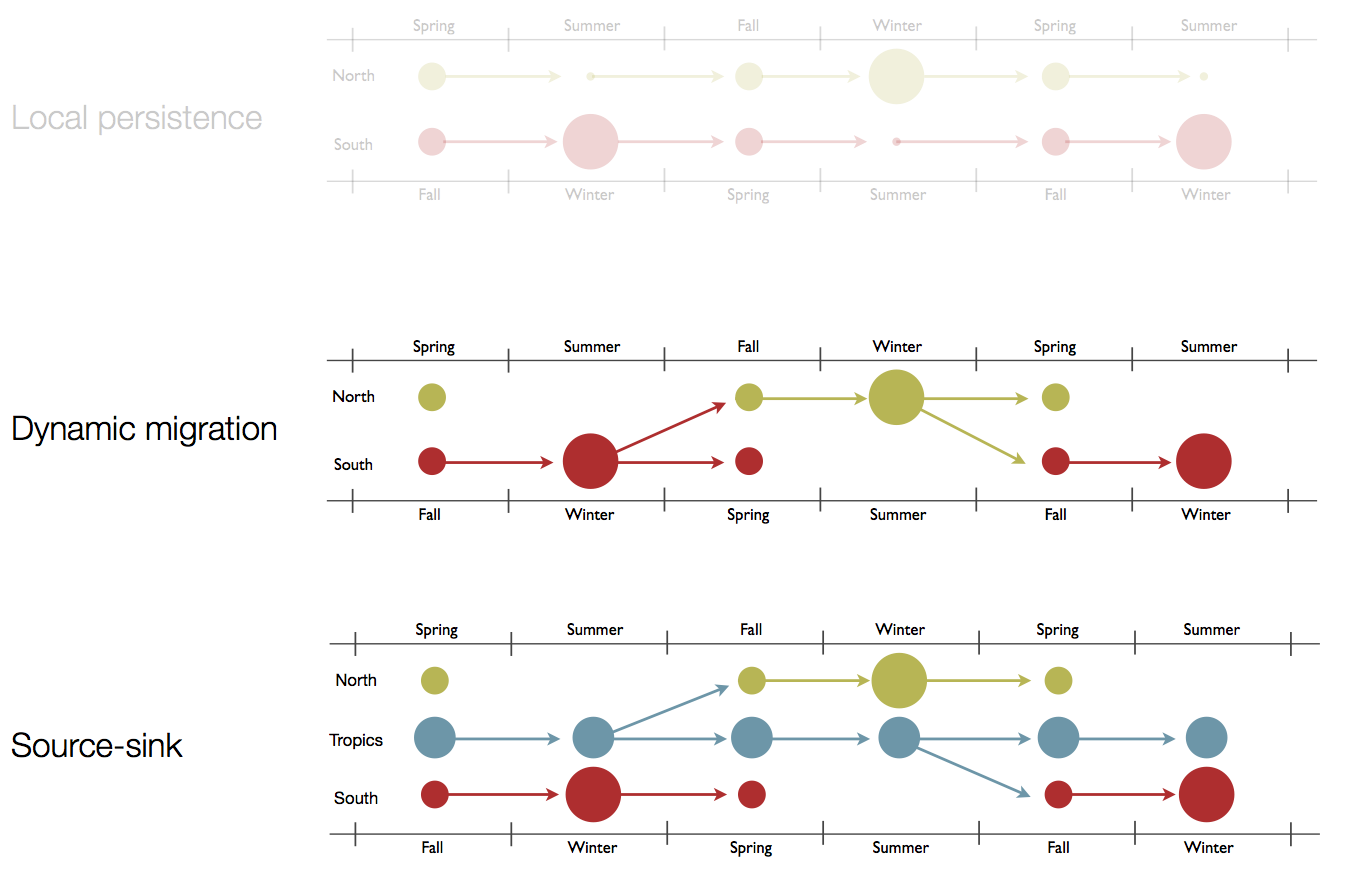
Hypotheses of influenza circulation patterns
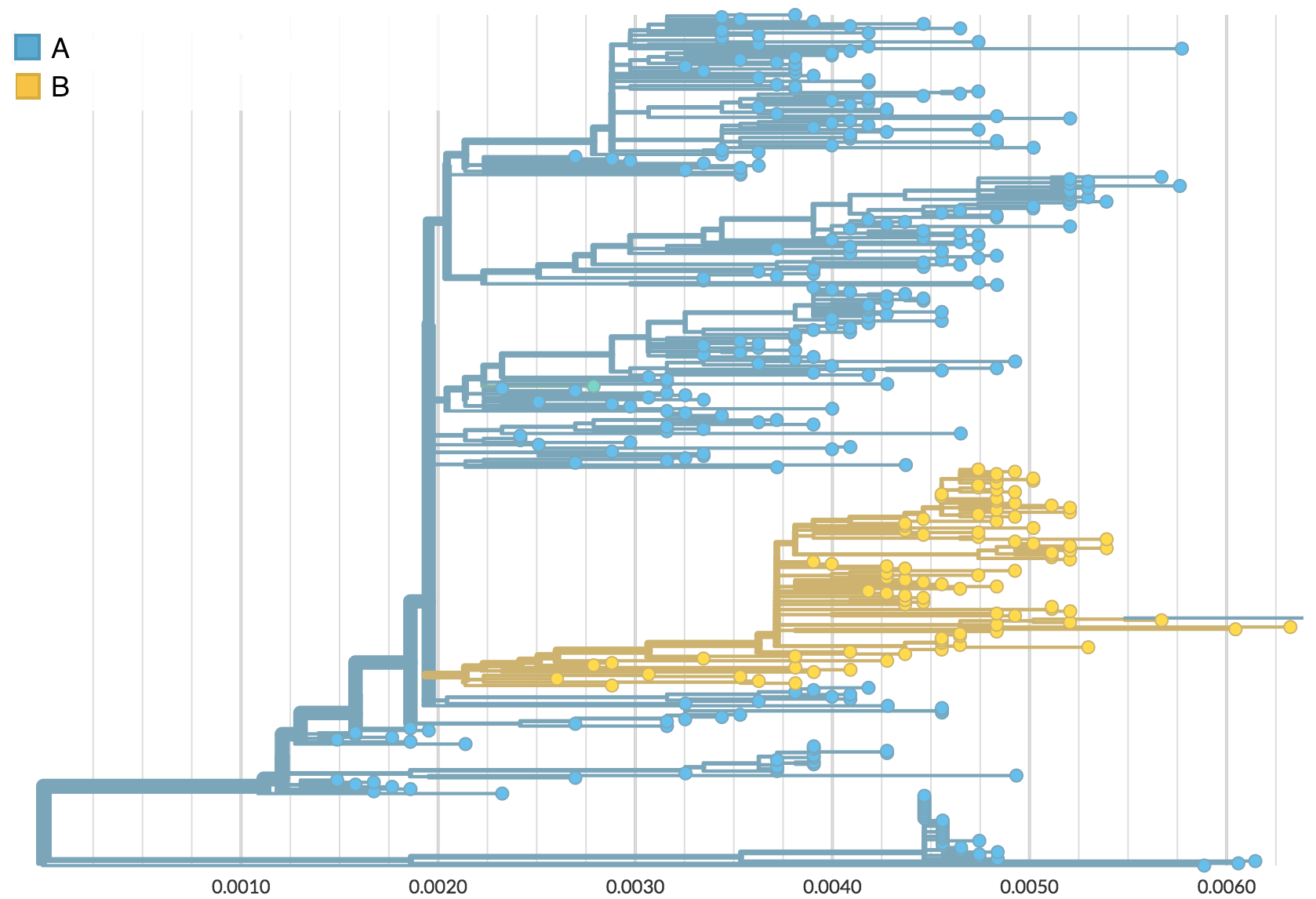
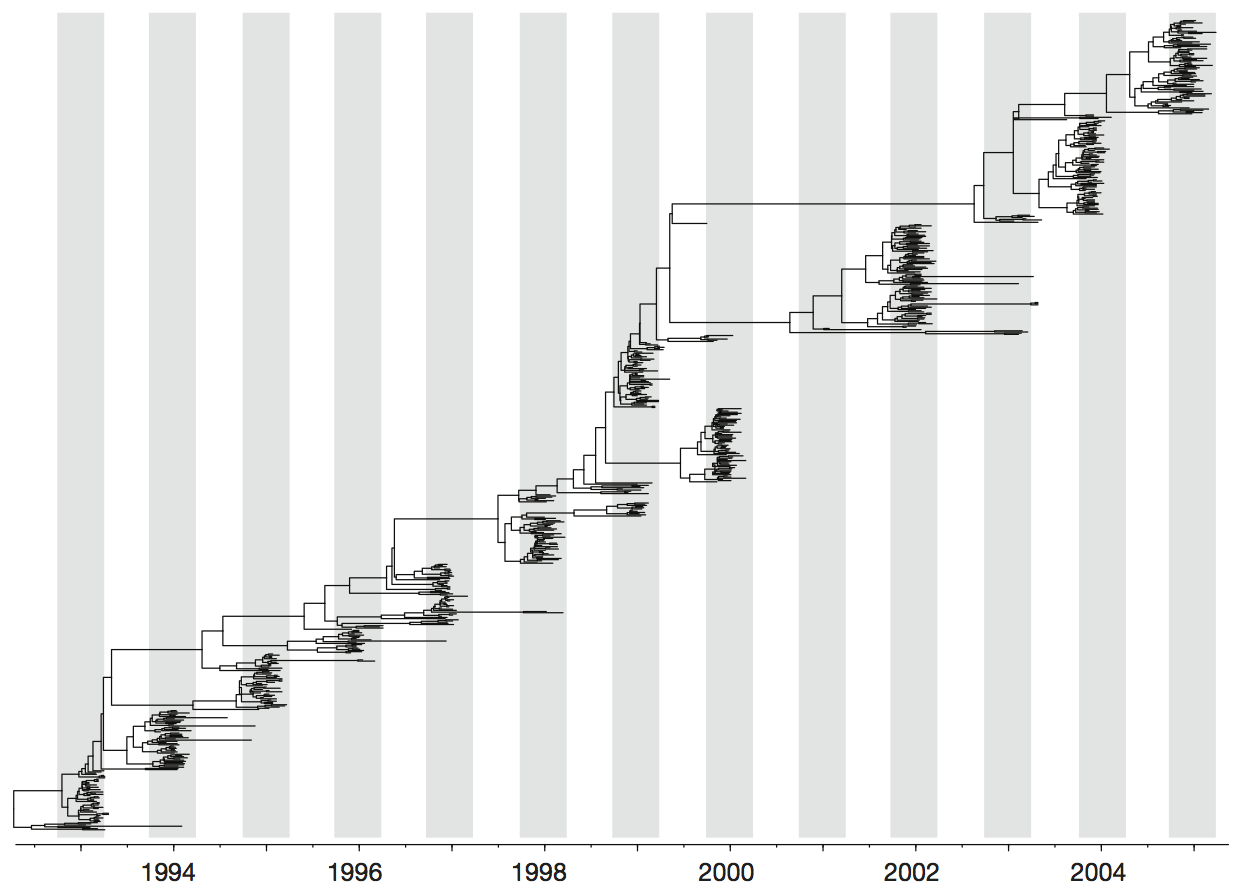
Influenza H3 genealogy for NY state viruses
Hypotheses of influenza circulation patterns
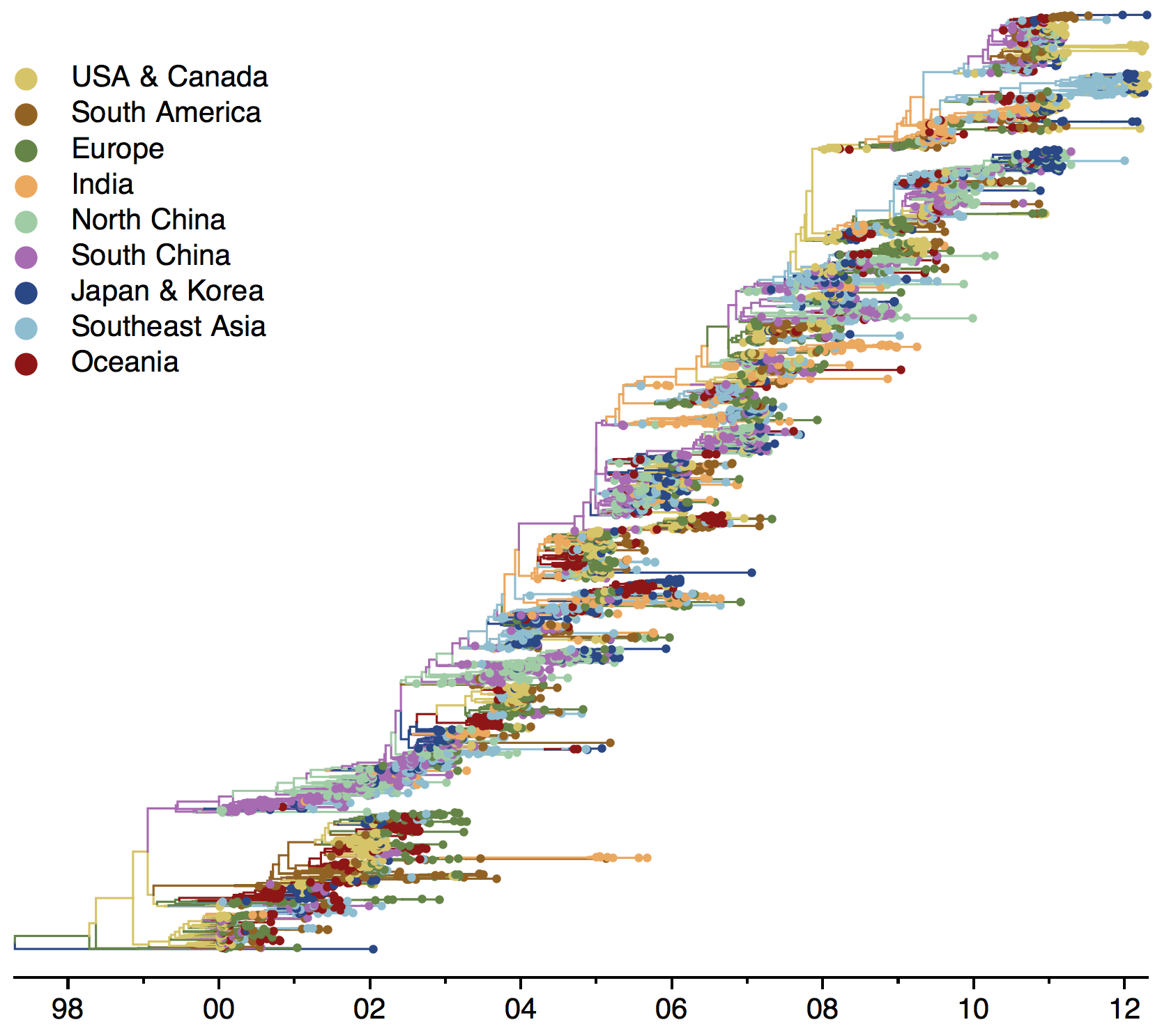
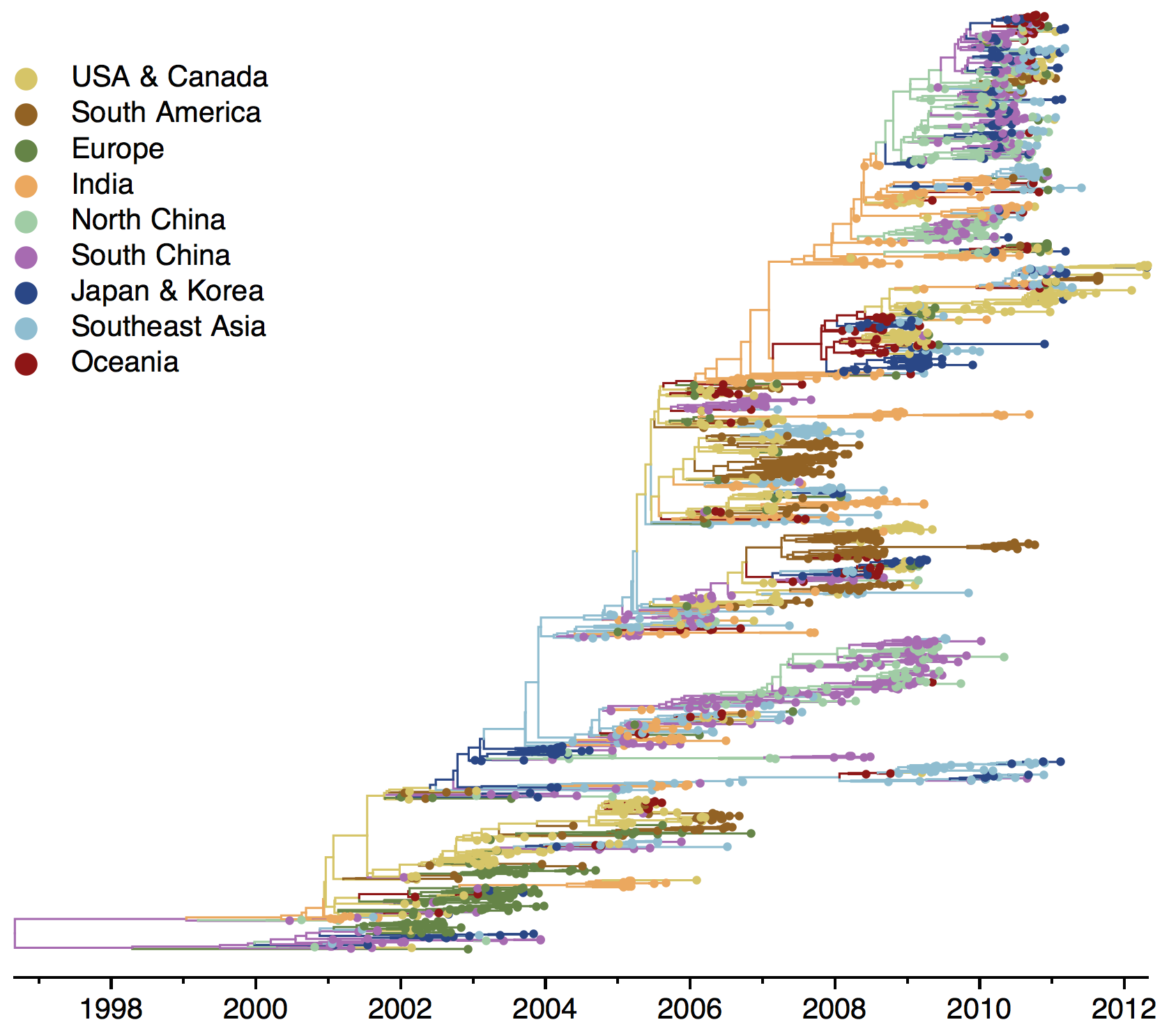
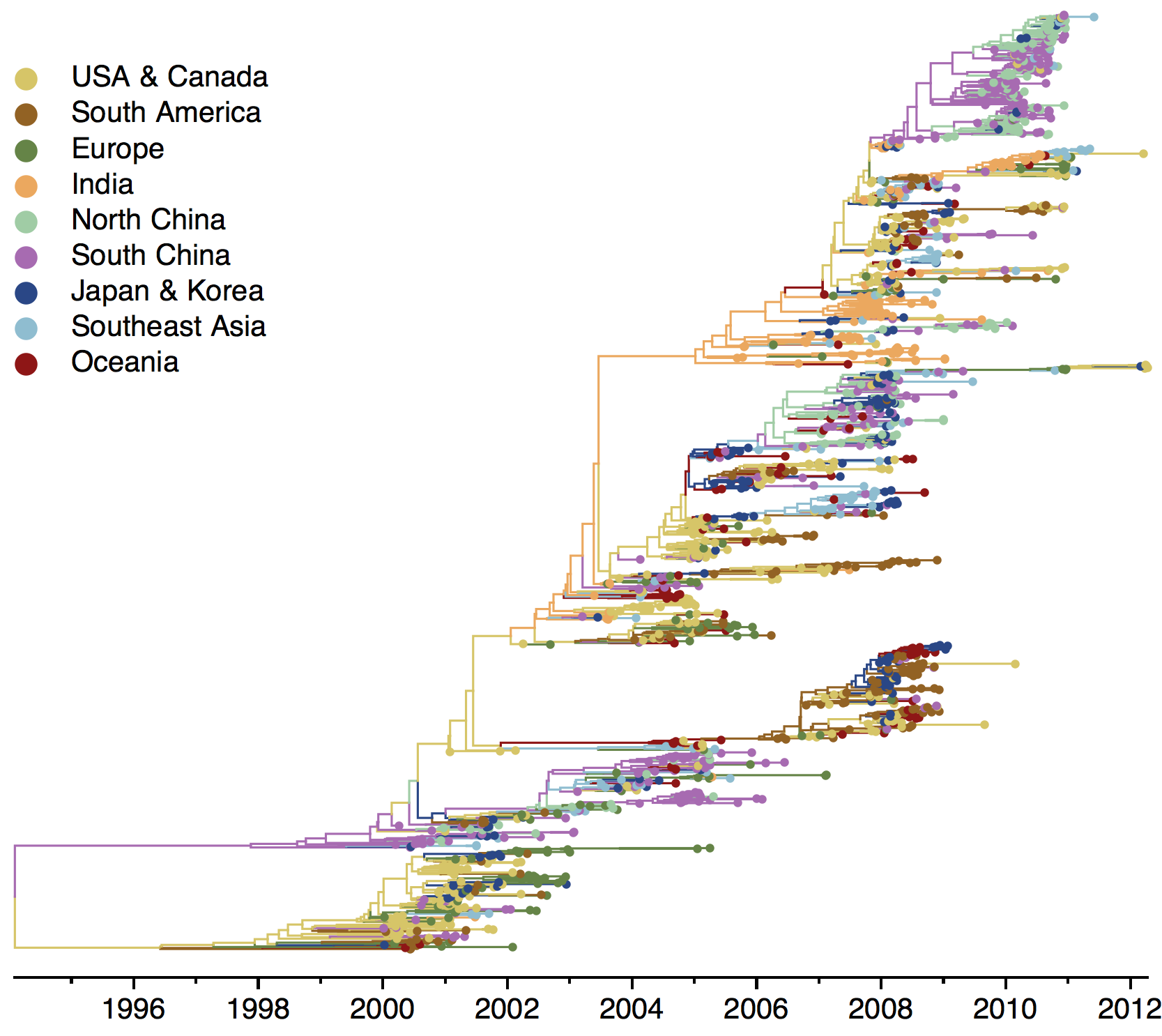
Sample H3N2 from around the world
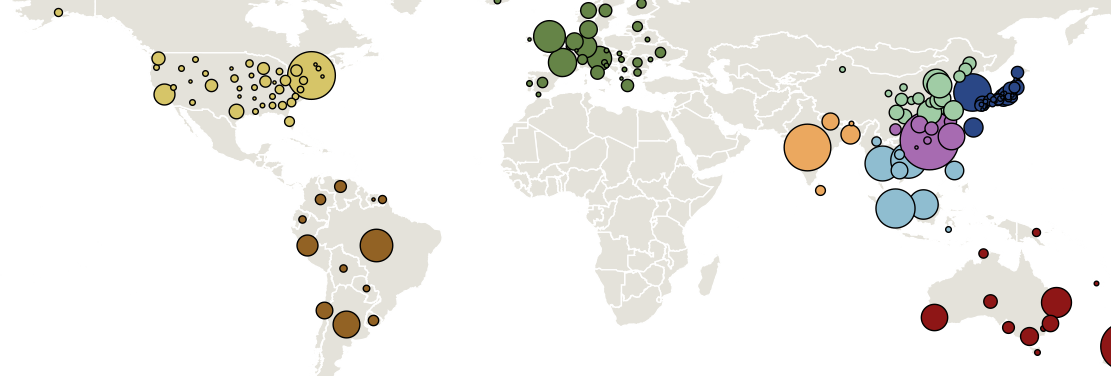
Treating geographic state as an evolving character
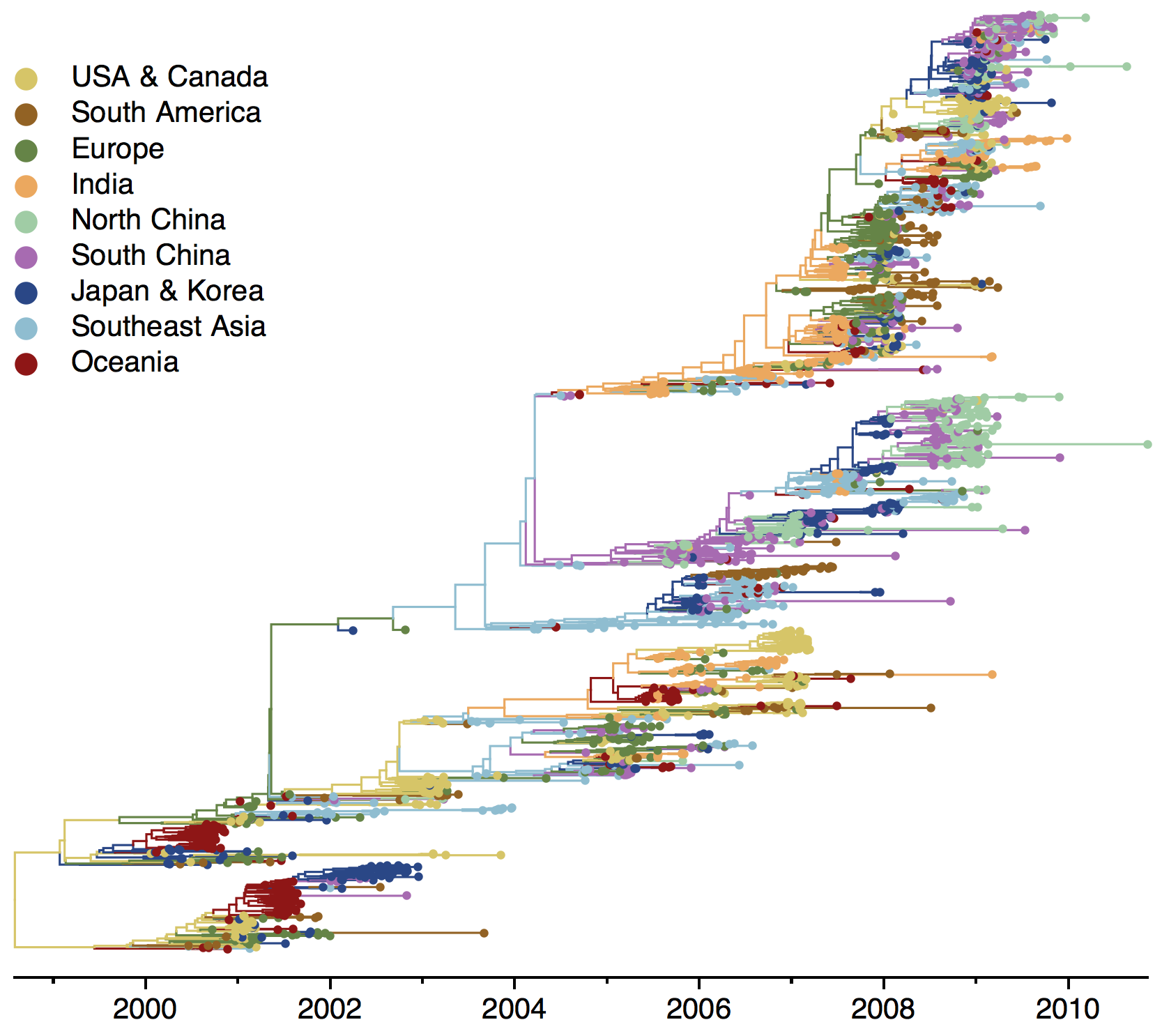
Phylogeny of H3 with geographic history
Infer geographic transition matrix
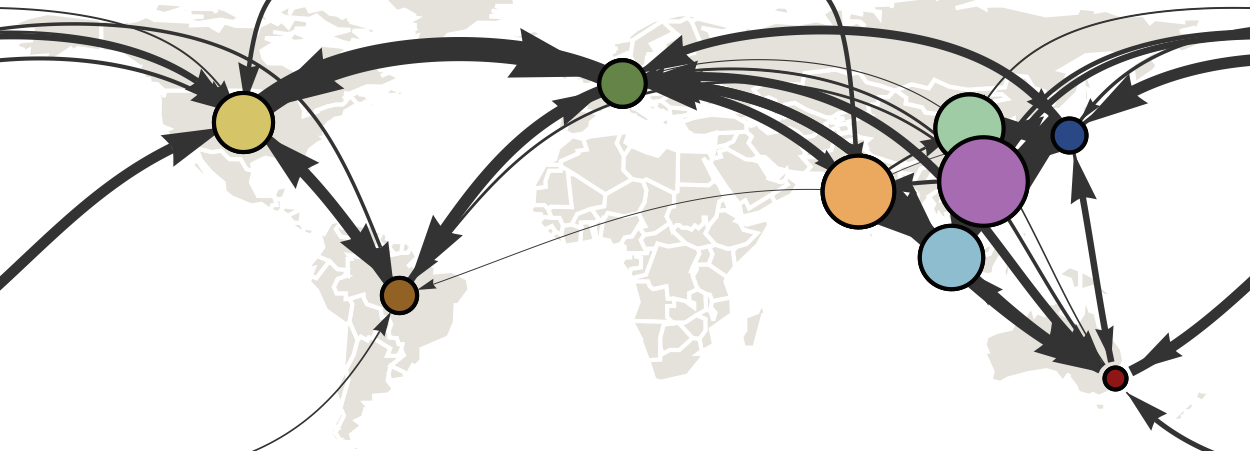
Air travel predicts migration rates
Geographic location of phylogeny trunk
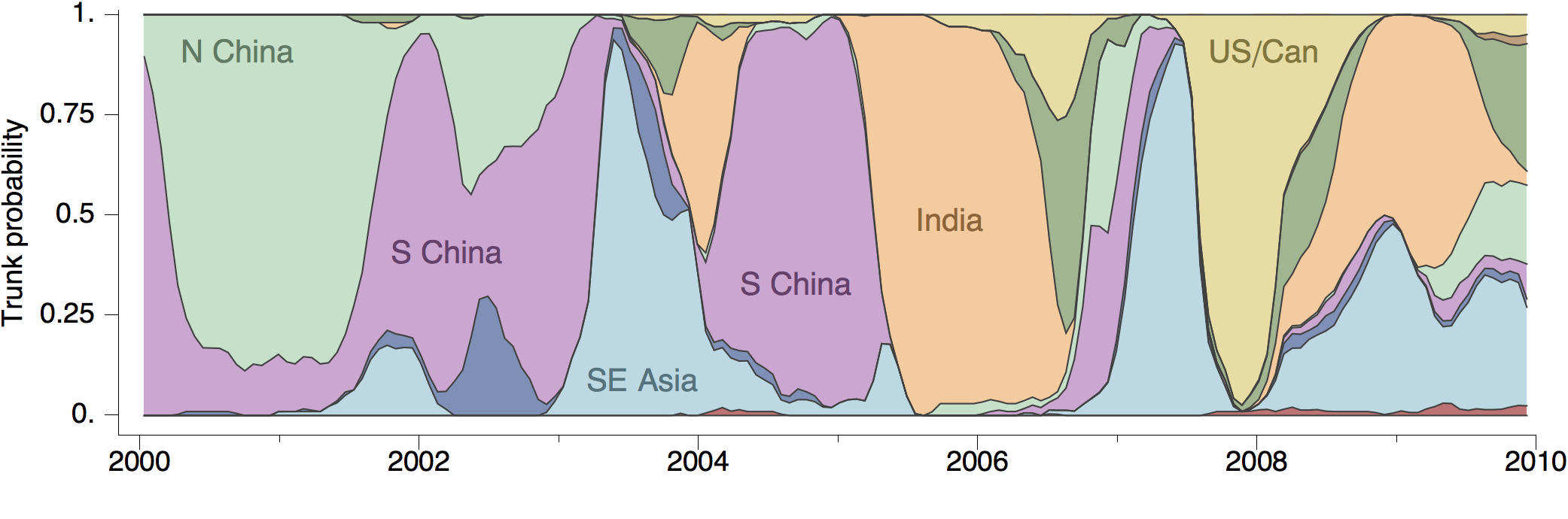
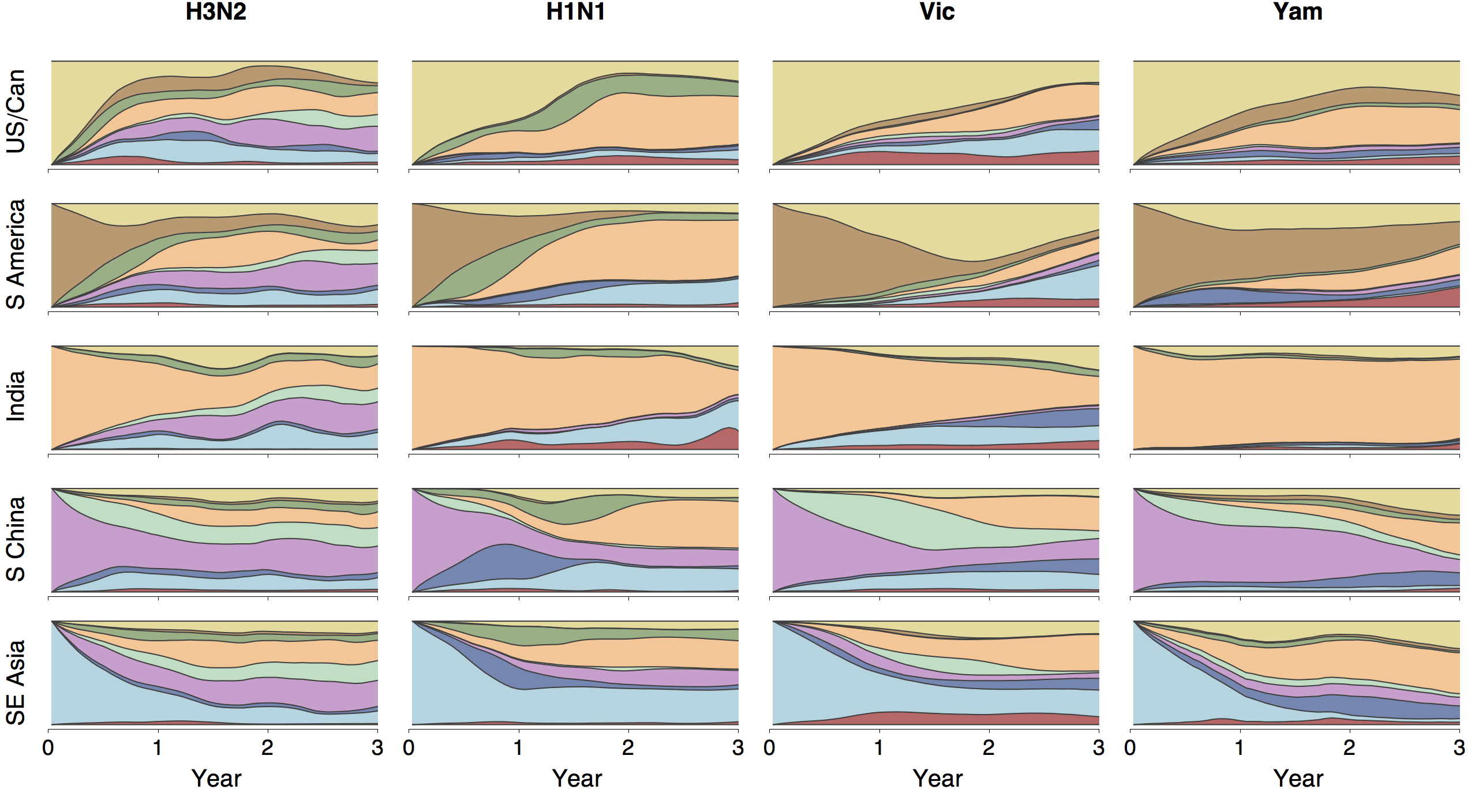
Region-specific ancestry
Phylogenies across subtypes / lineages
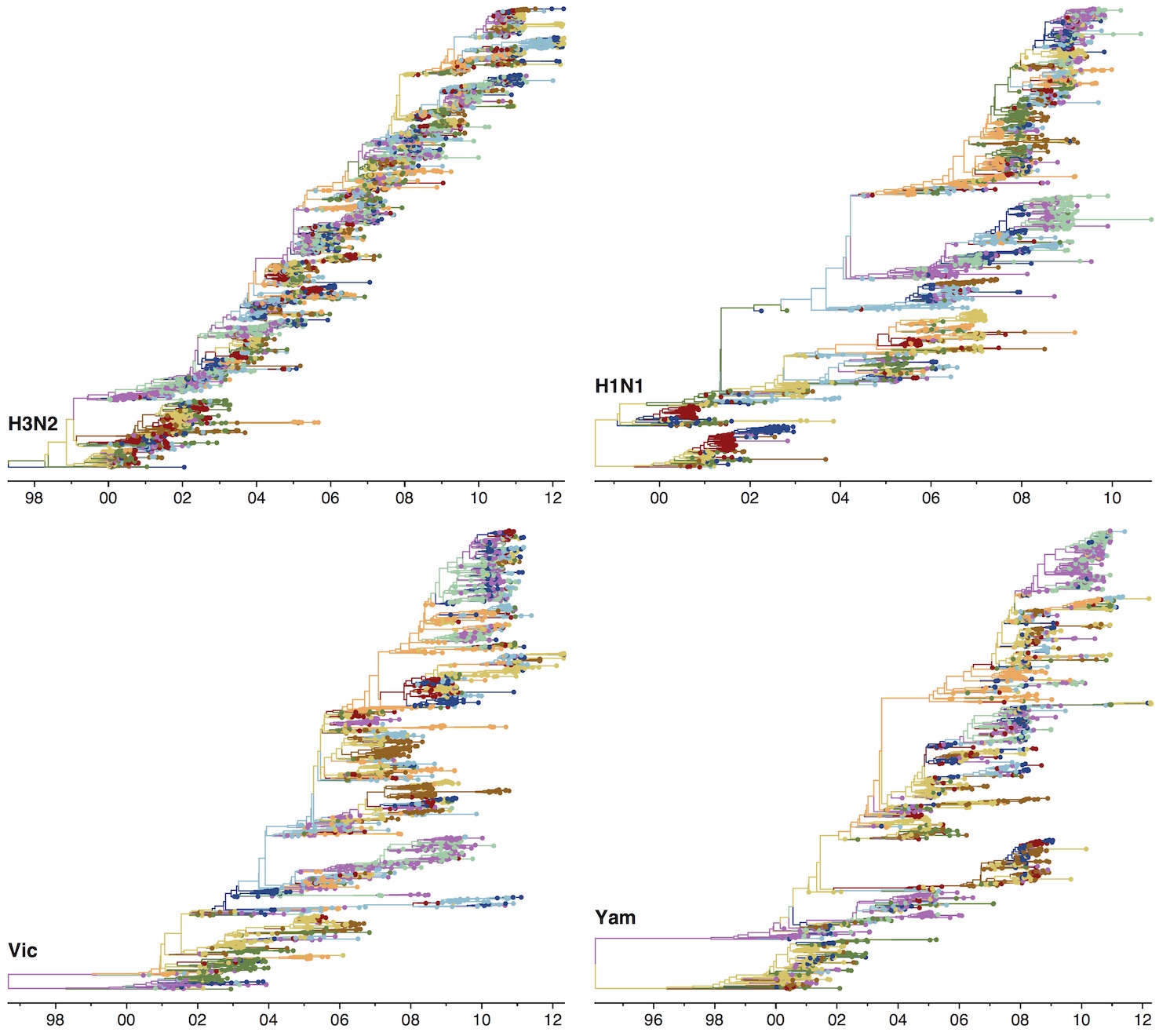
H3N2 phylogeny

H1N1 phylogeny
B/Vic phylogeny
B/Yam phylogeny
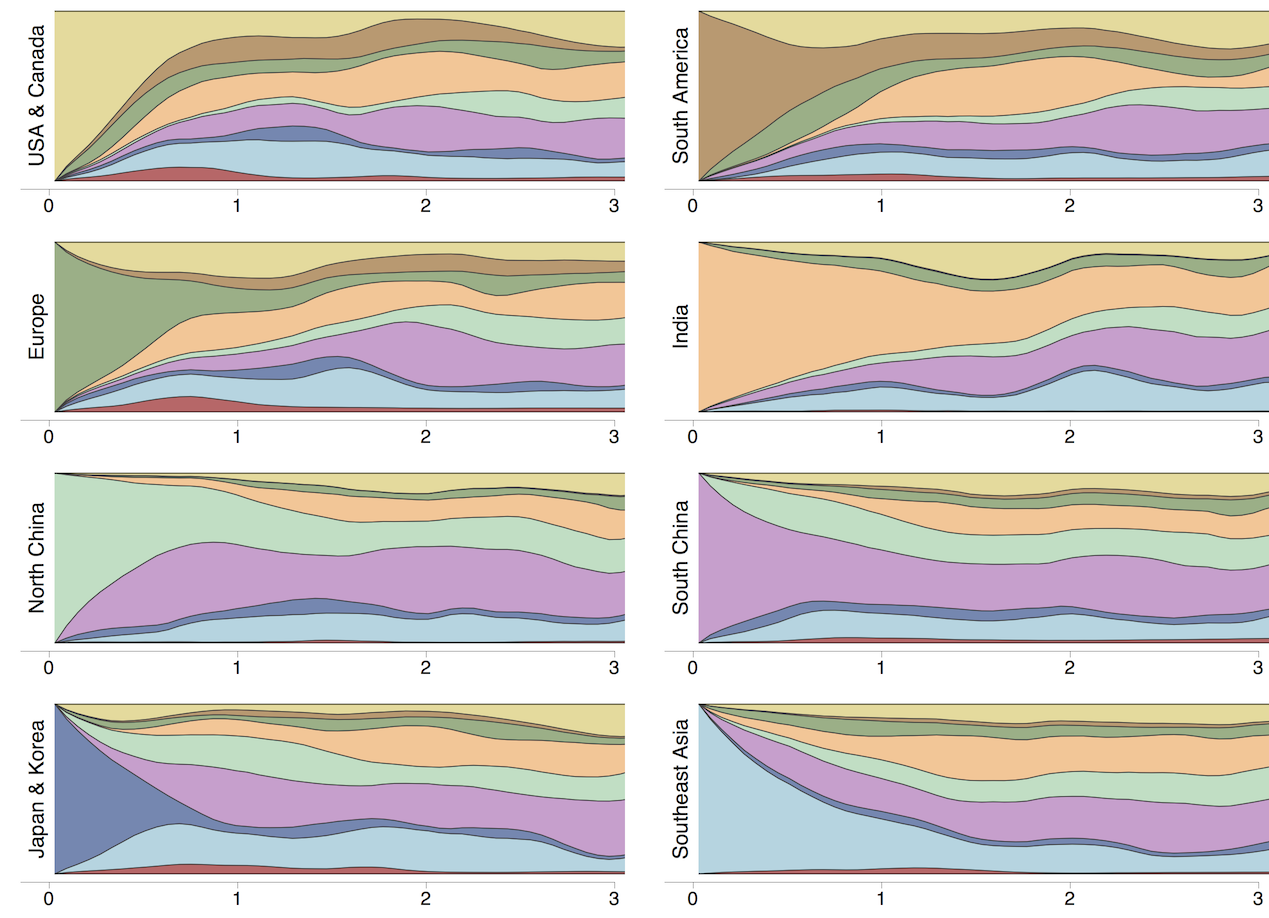
Ancestry patterns across lineages
Regional persistence patterns
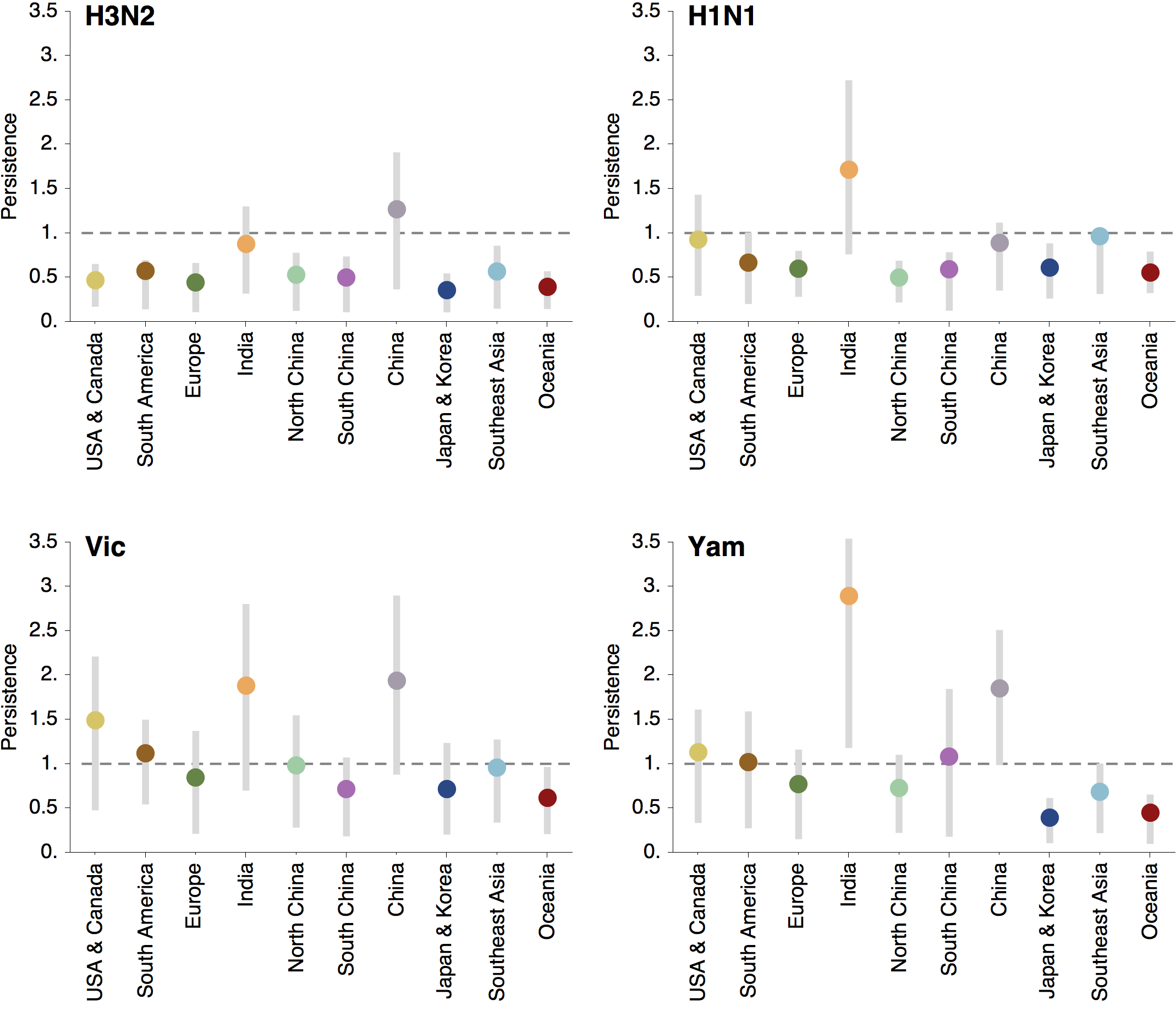
How to explain these differences?
Age distribution across viruses
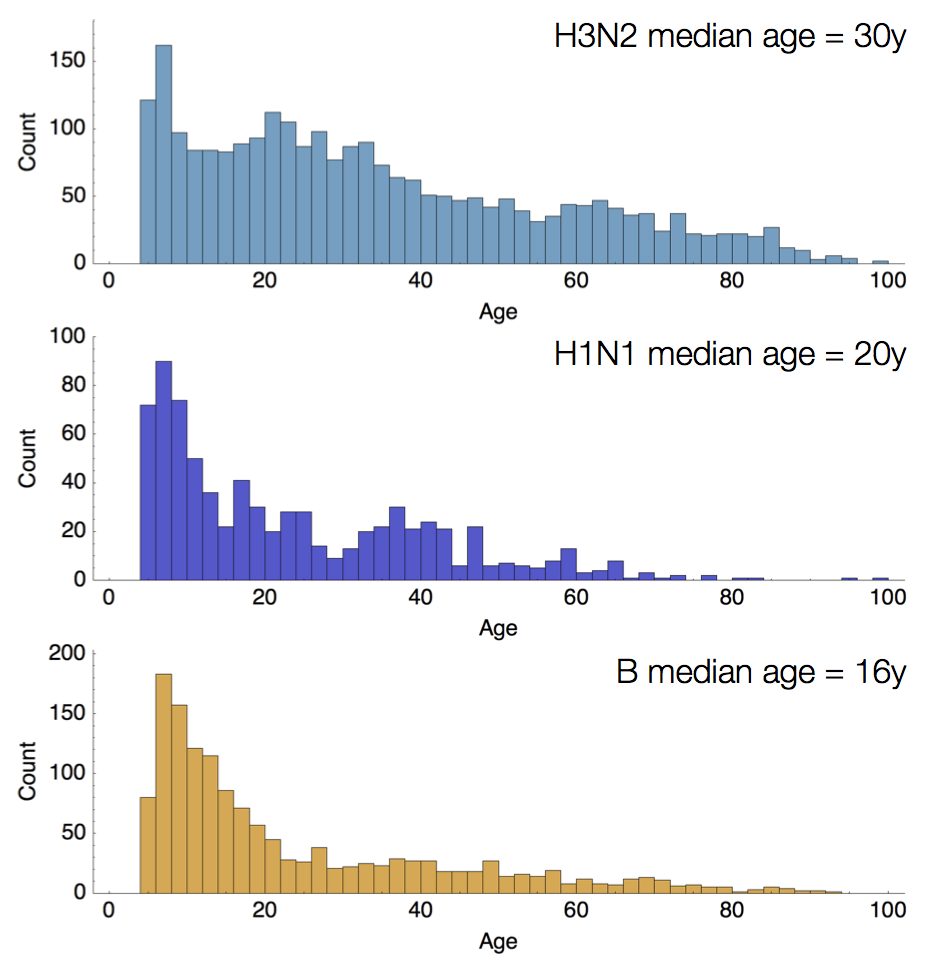
Air travel differences between adults and children
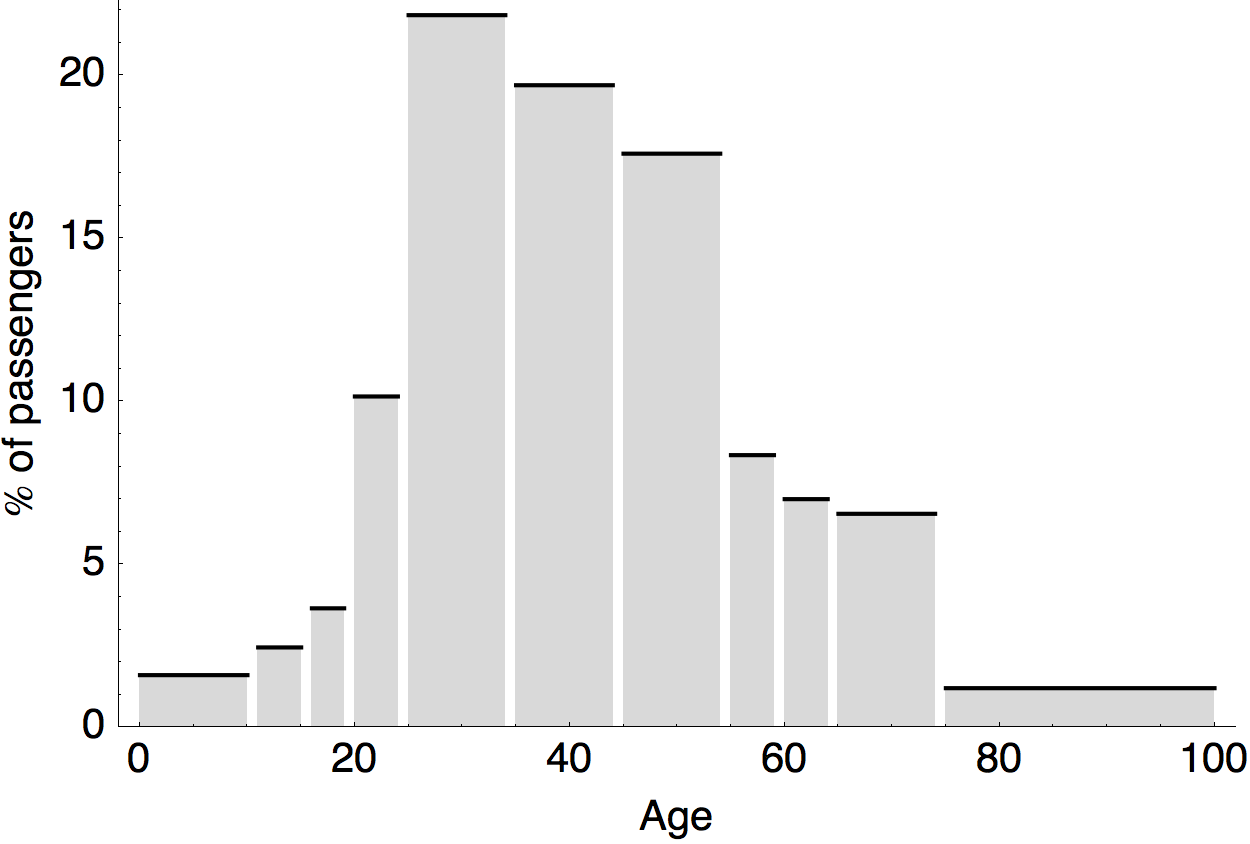
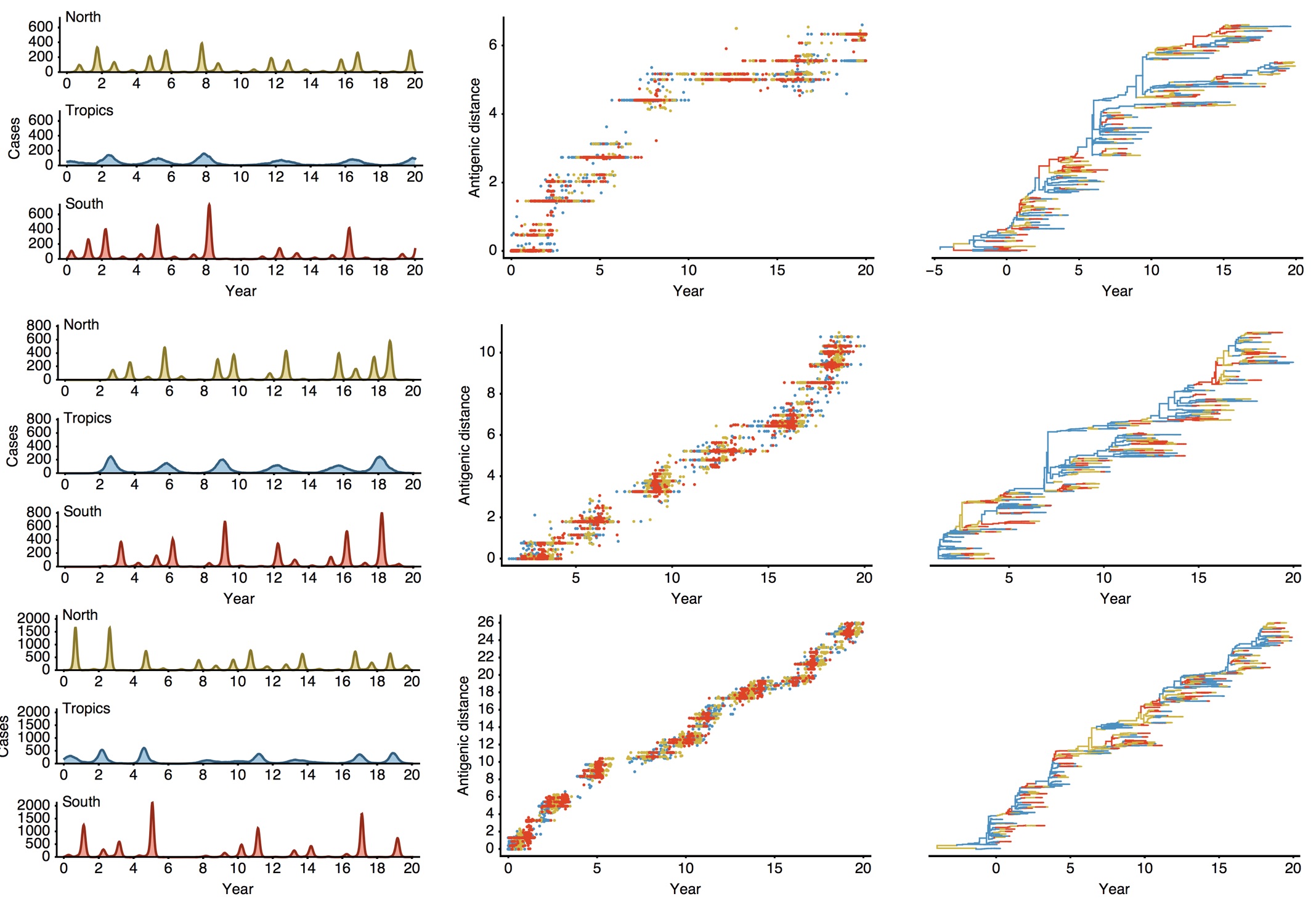
Epidemiological model of varying rates of antigenic drift
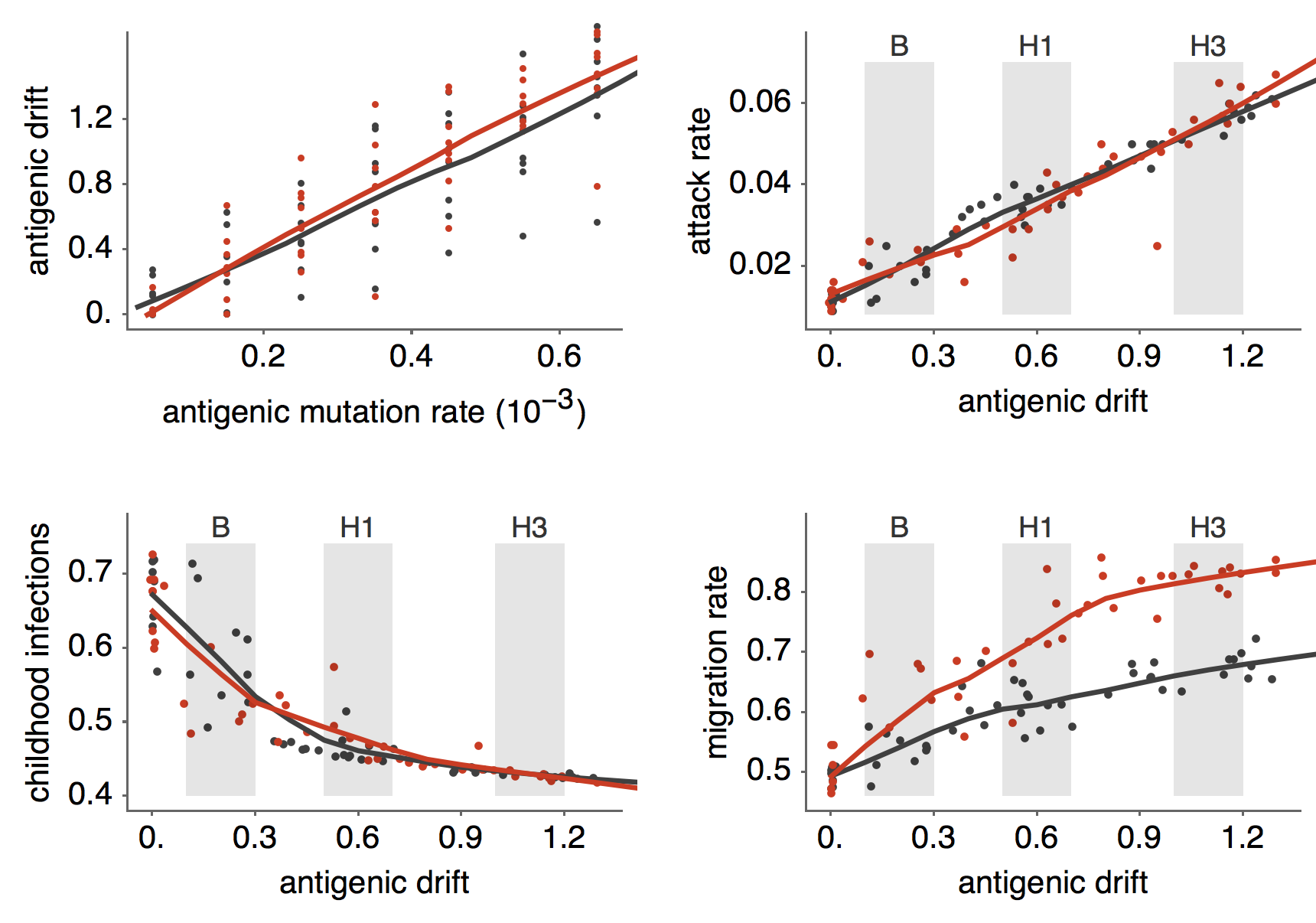
Results of varying antigenic drift
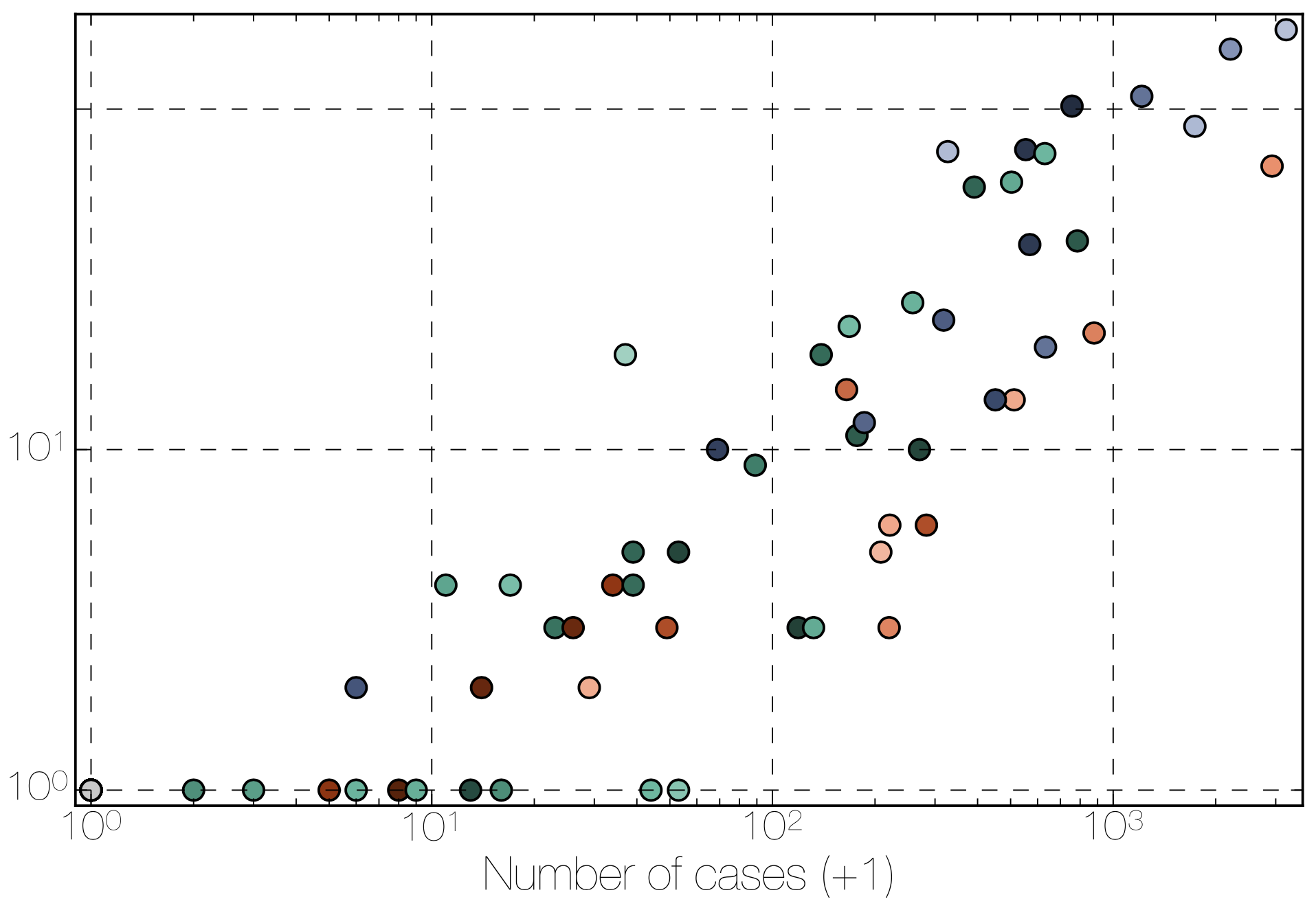
Sequencing of 1610 Ebola virus genomes collected during the 2013-2016 West African epidemic
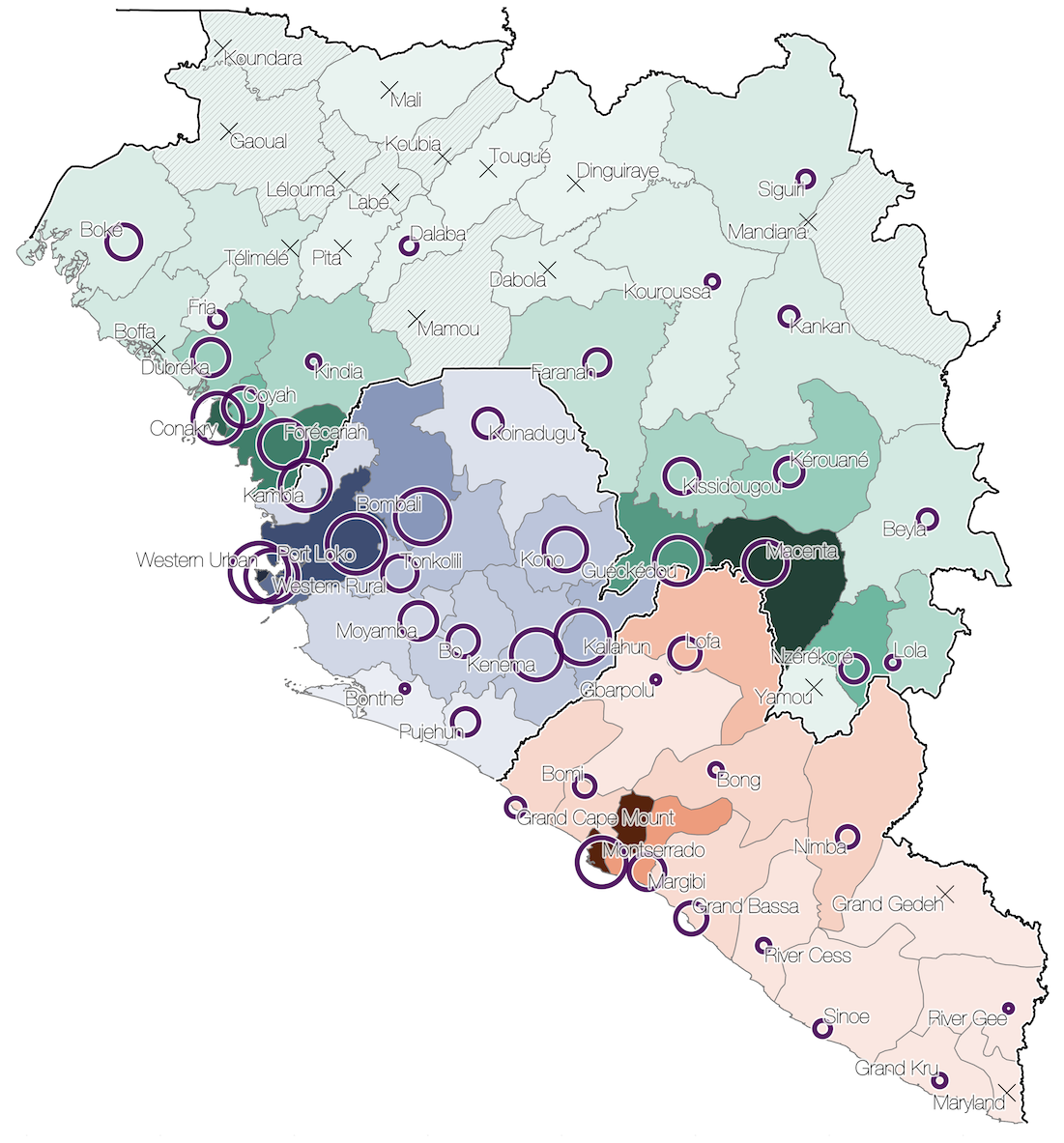
Sequenced genomes were representative of spatiotemporal diversity
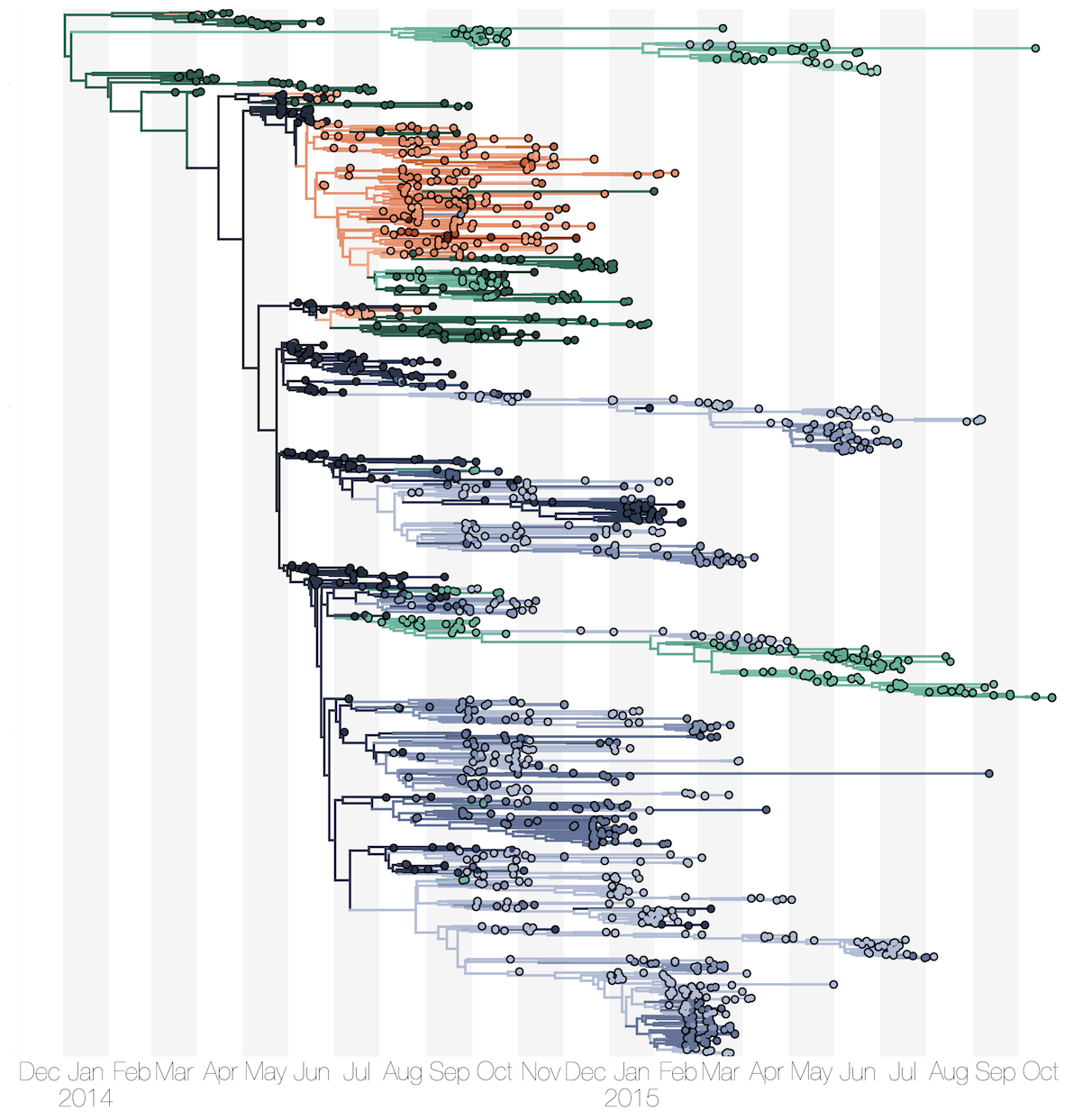
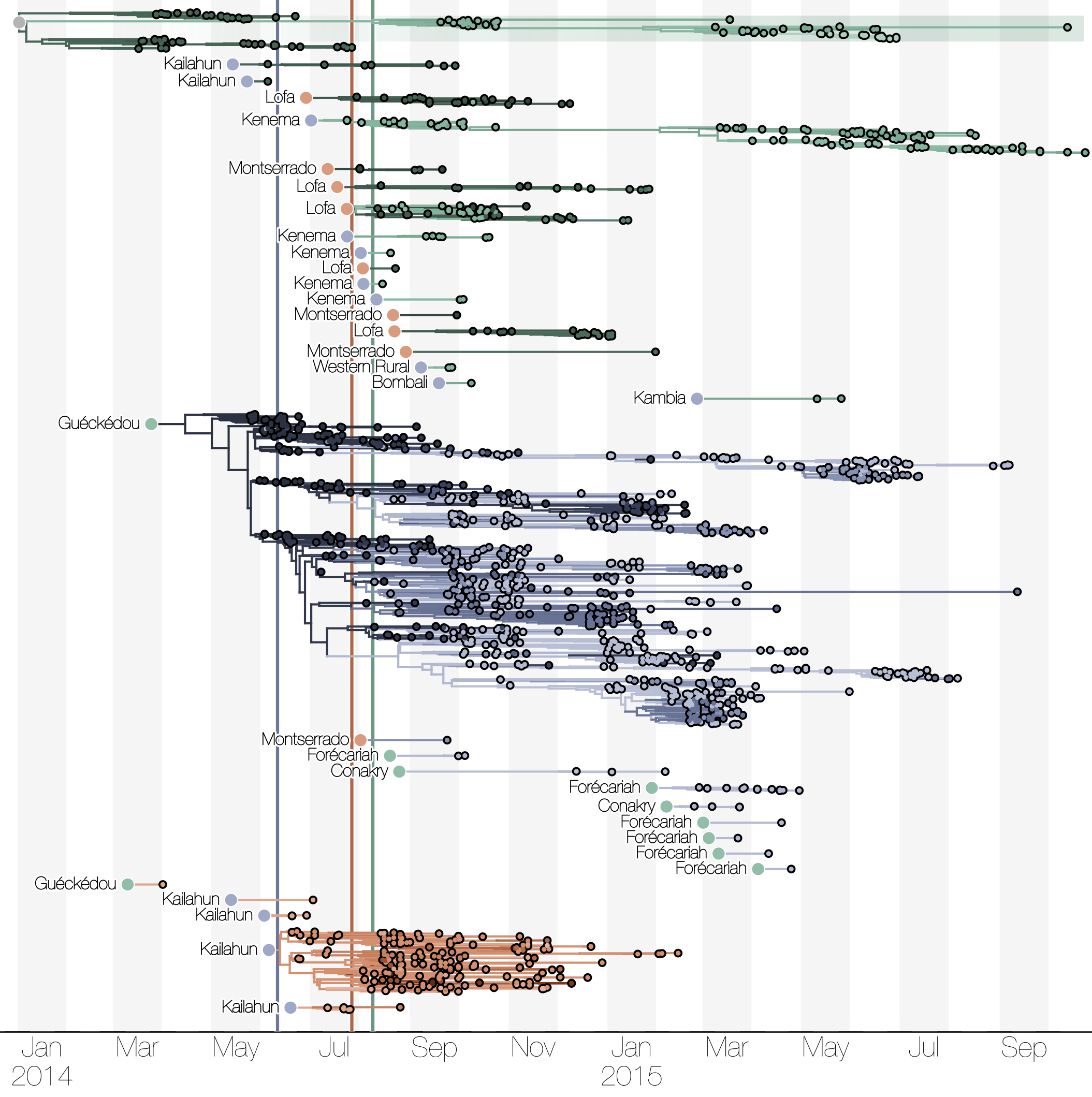
Phylogenetic reconstruction of epidemic
Tracking migration events
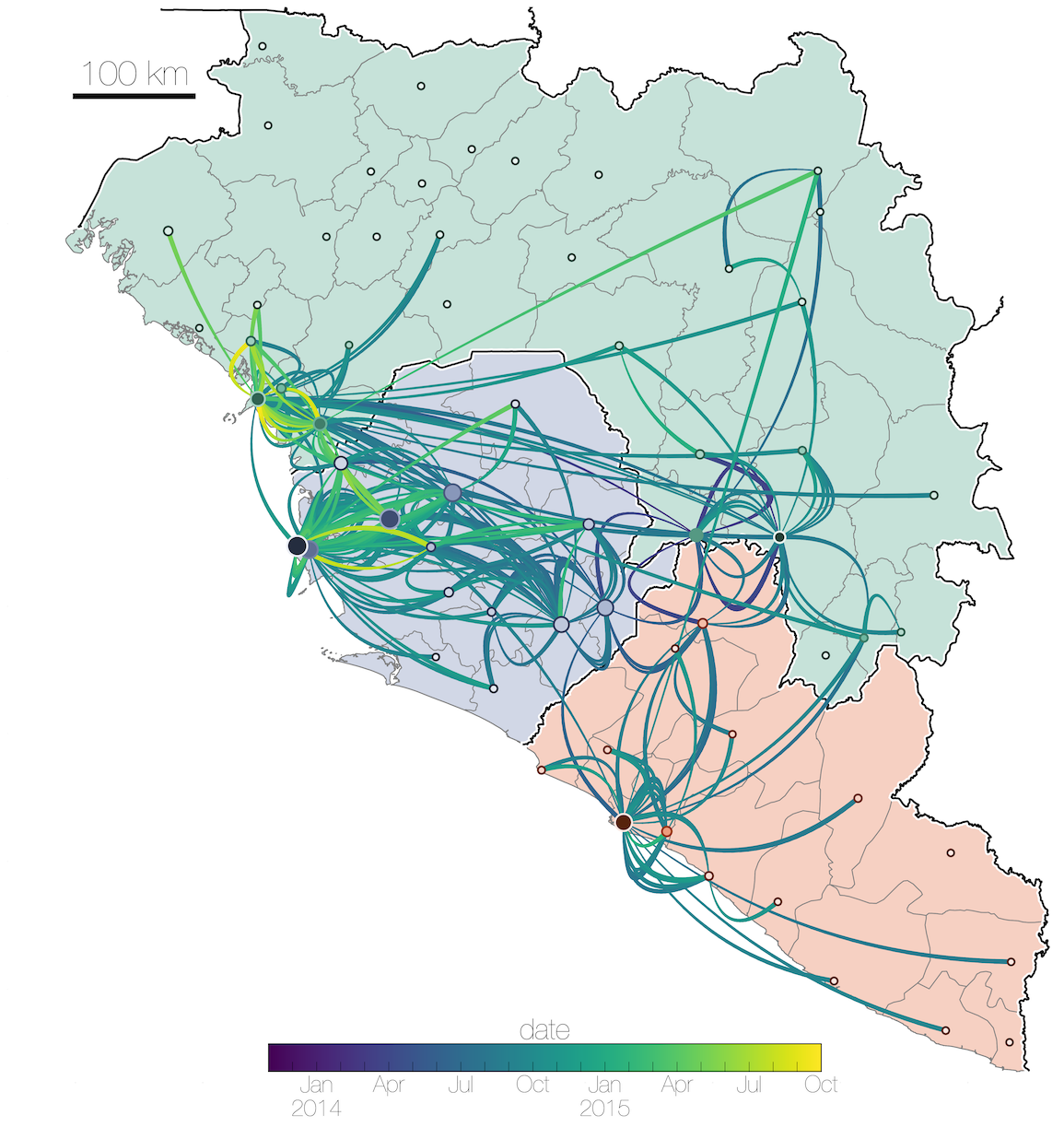
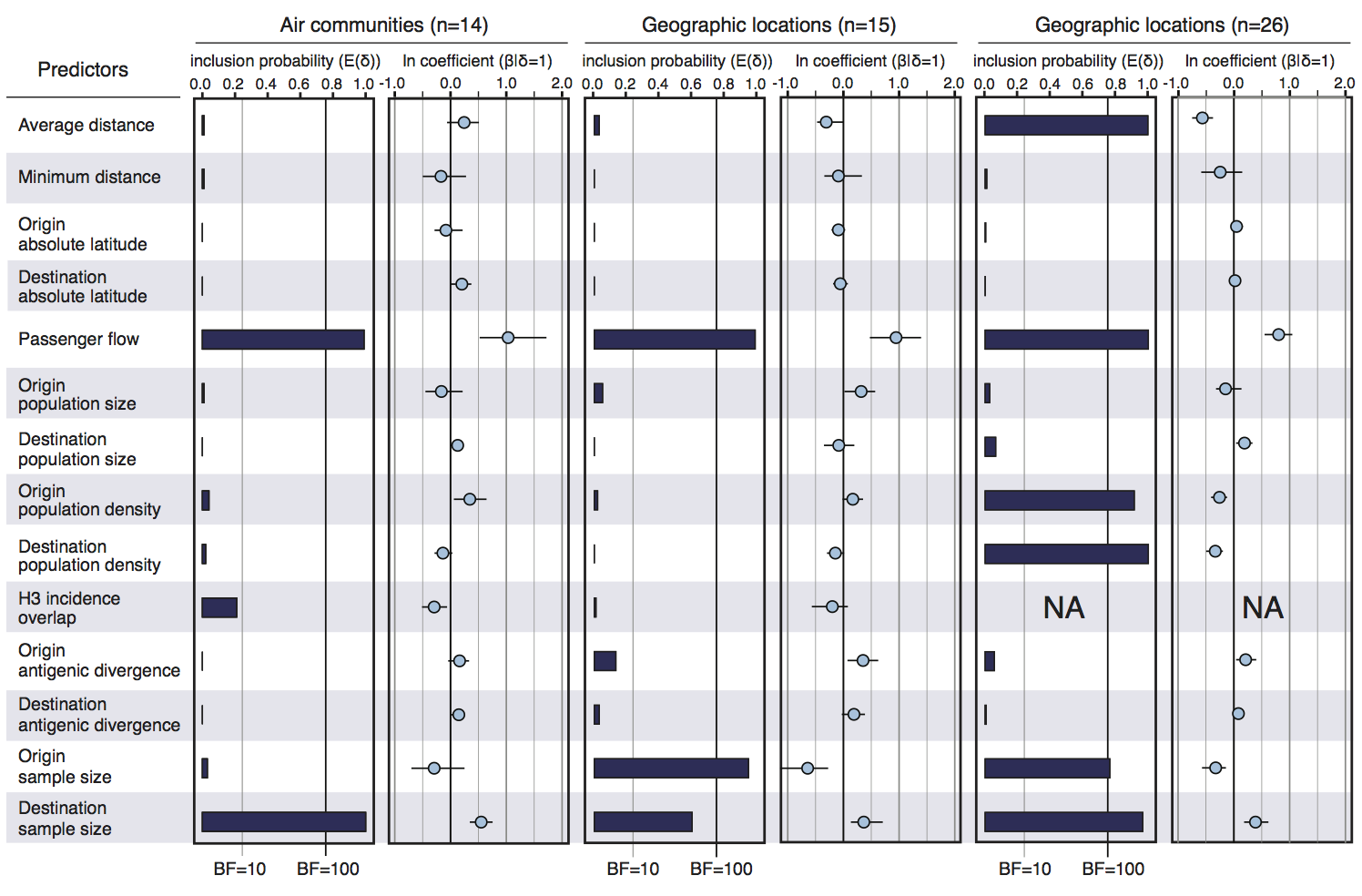
Factors influencing migration rates
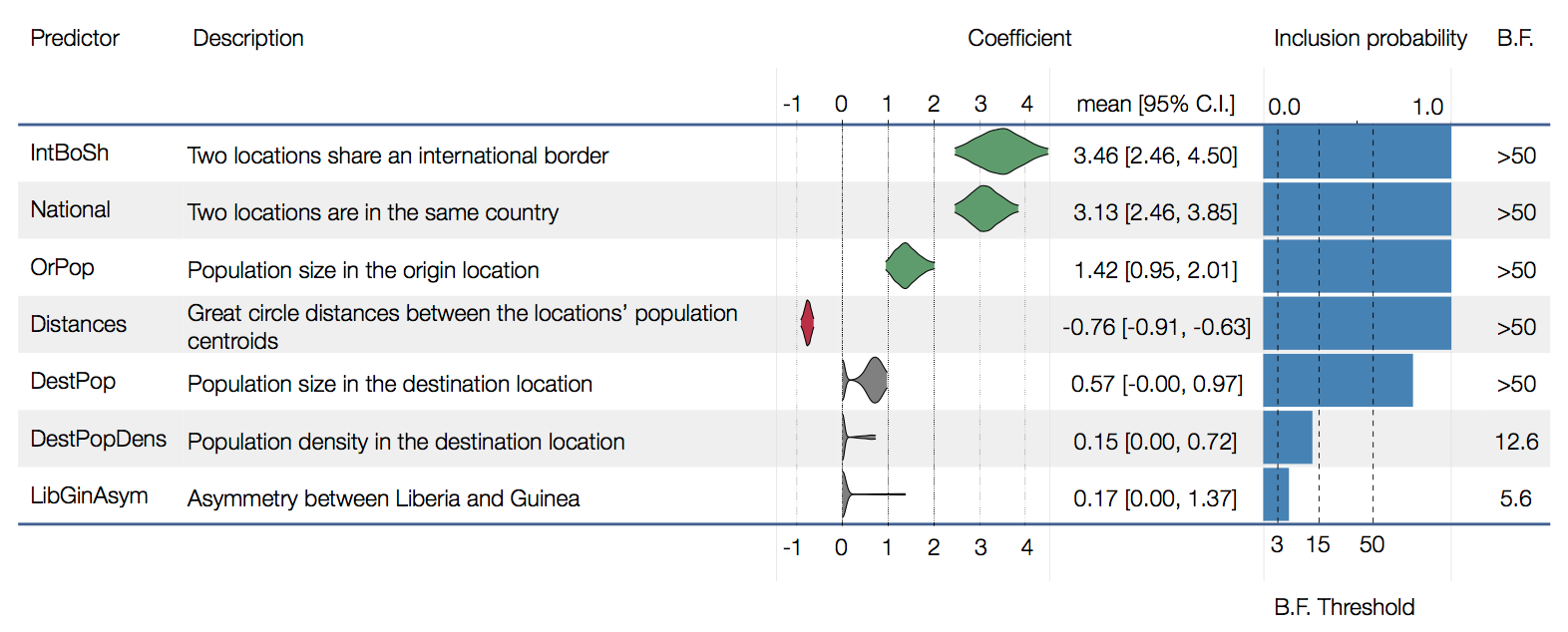
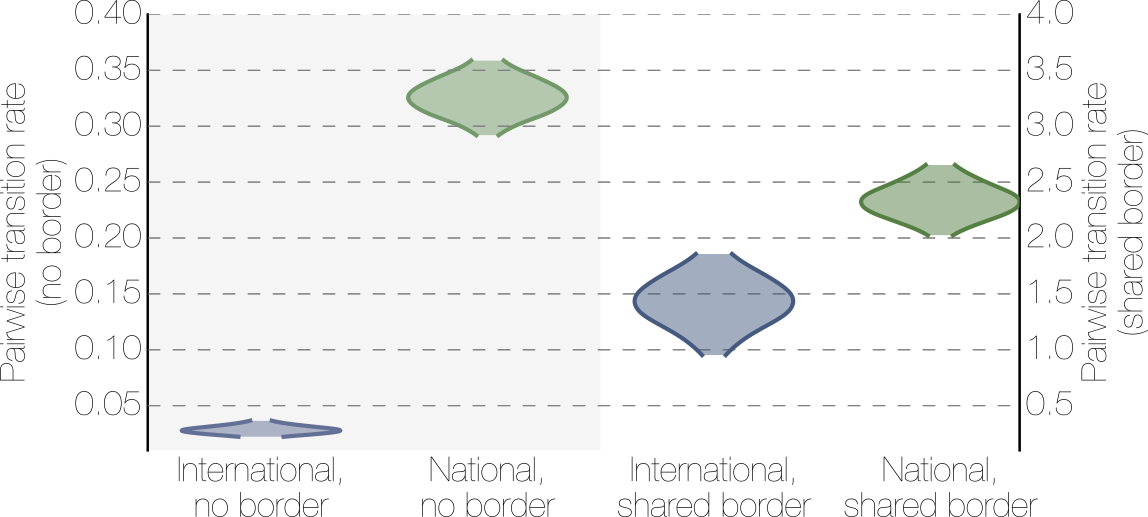
Effect of borders on migration rates
Spatial structure at the country level
Substantial mixing at the regional level
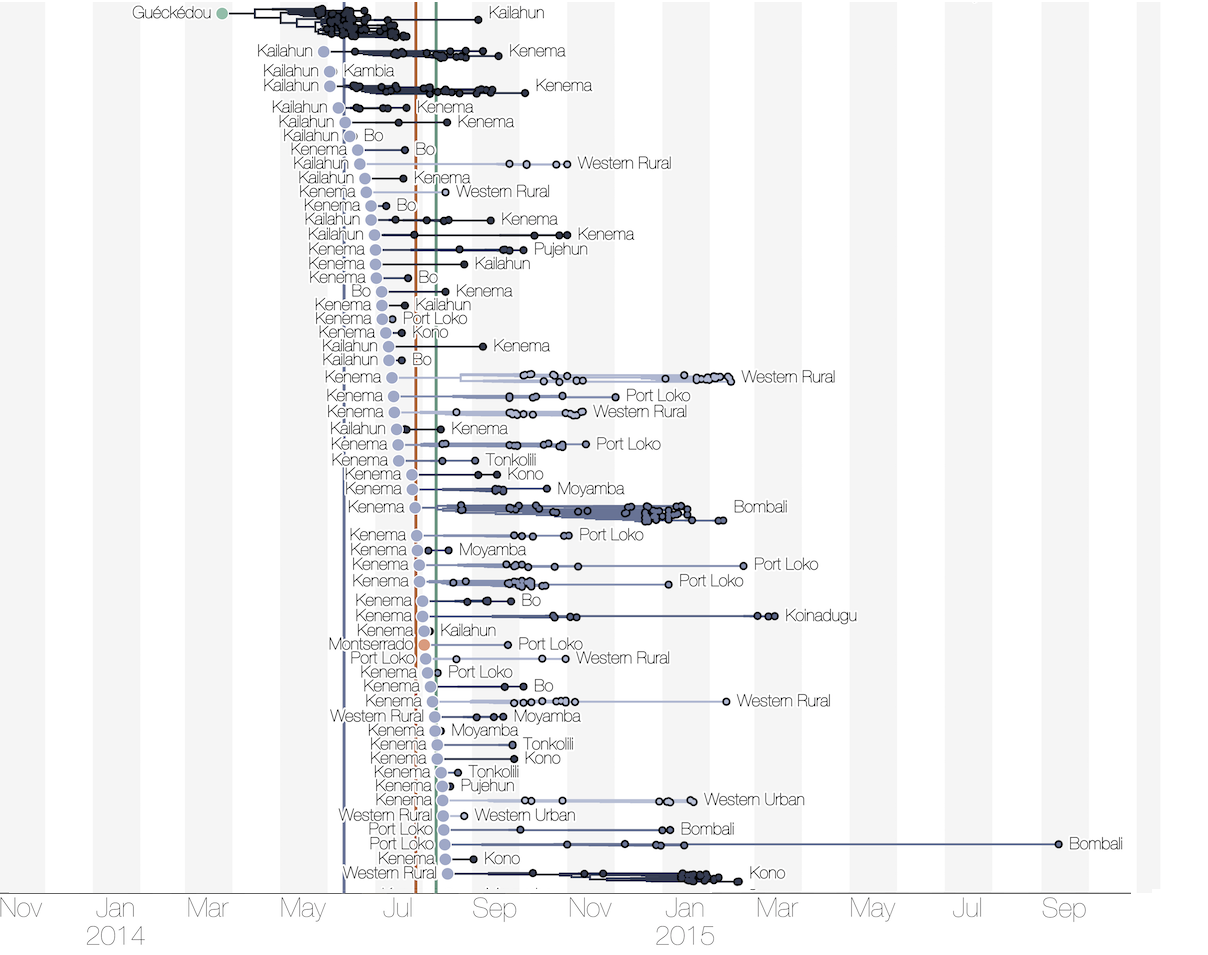
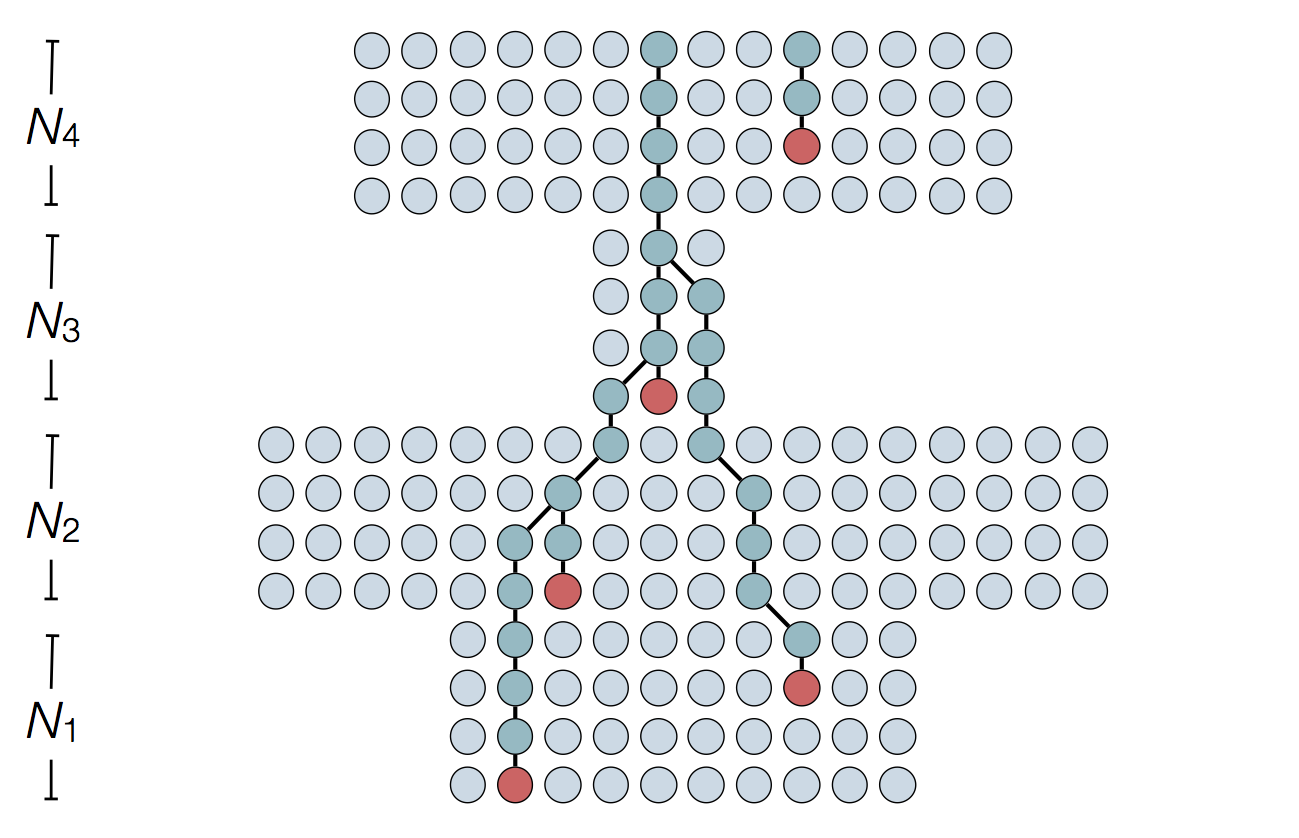
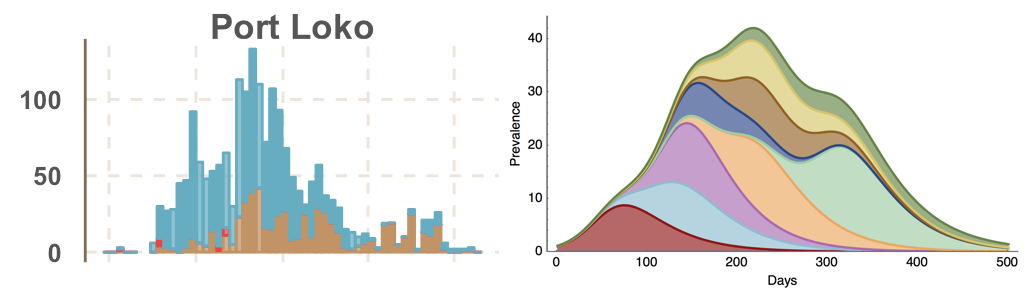
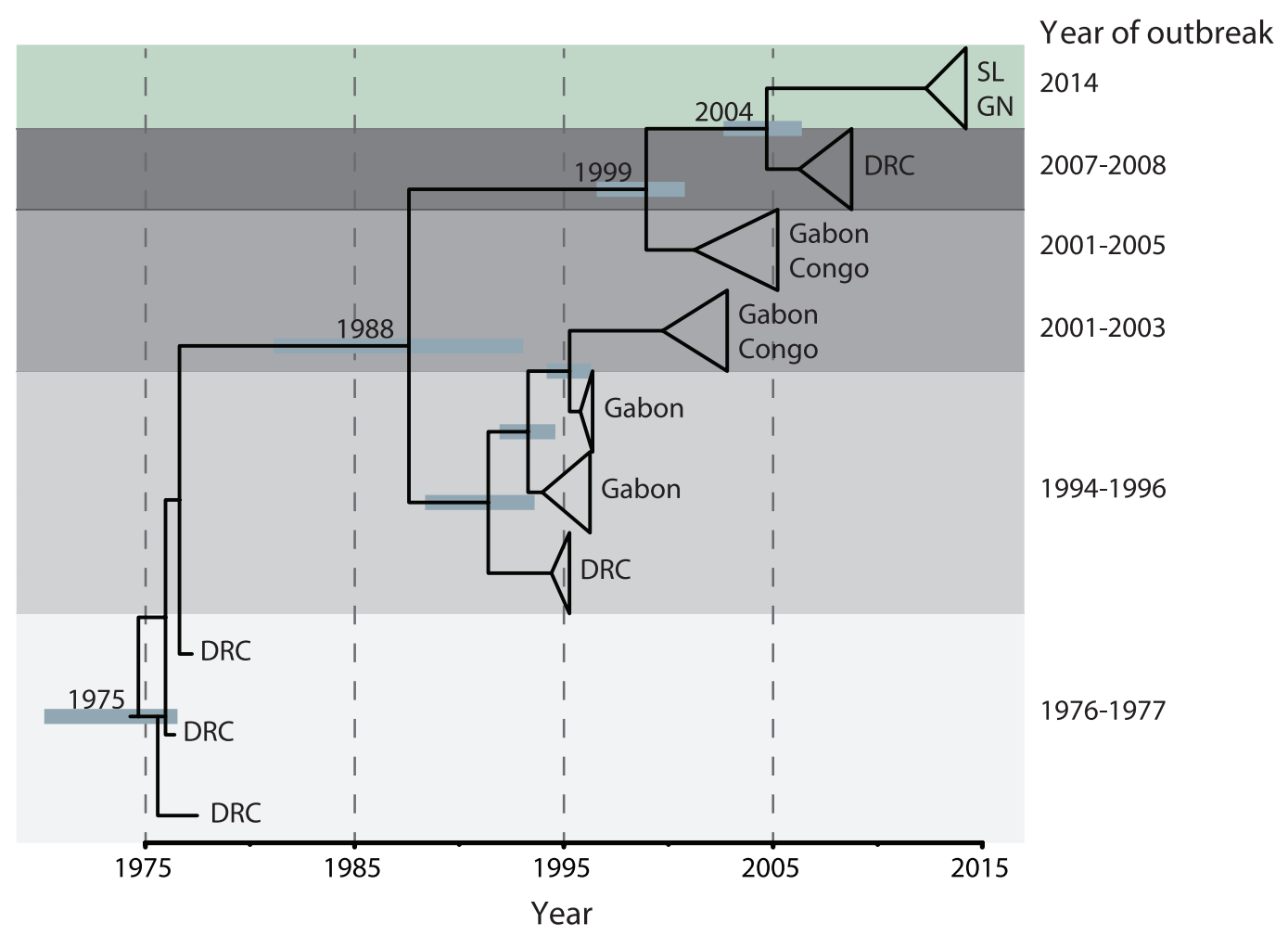
Each introduction results in a minor outbreak
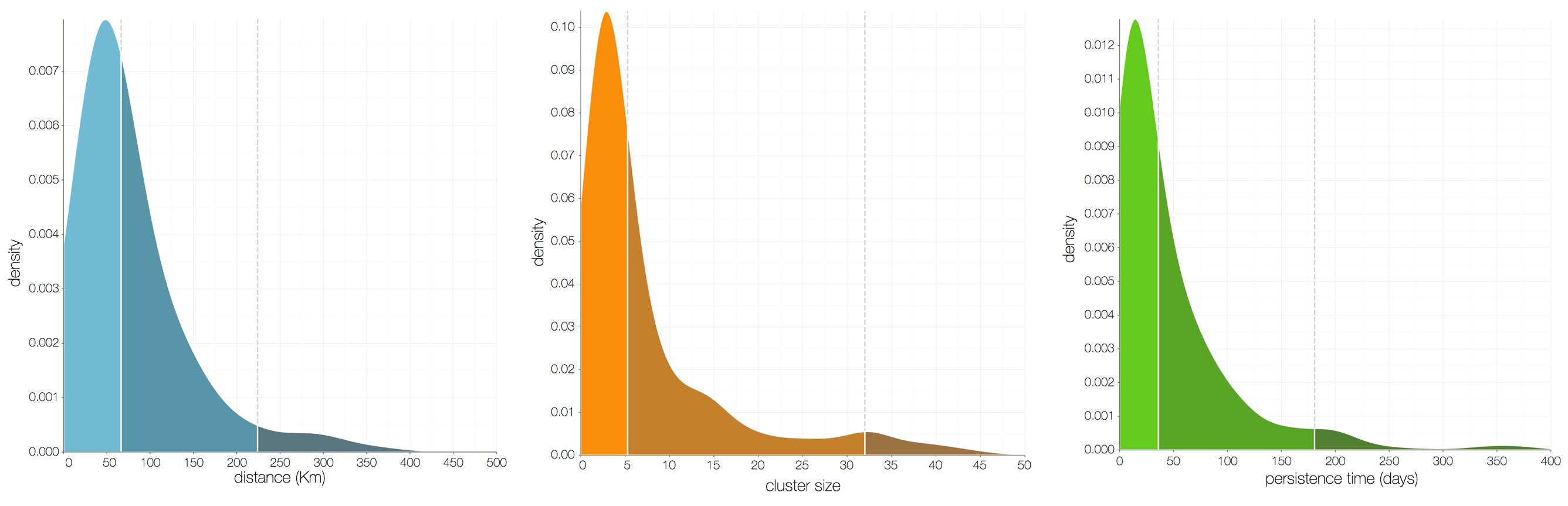
Regional outbreaks due to multiple introductions
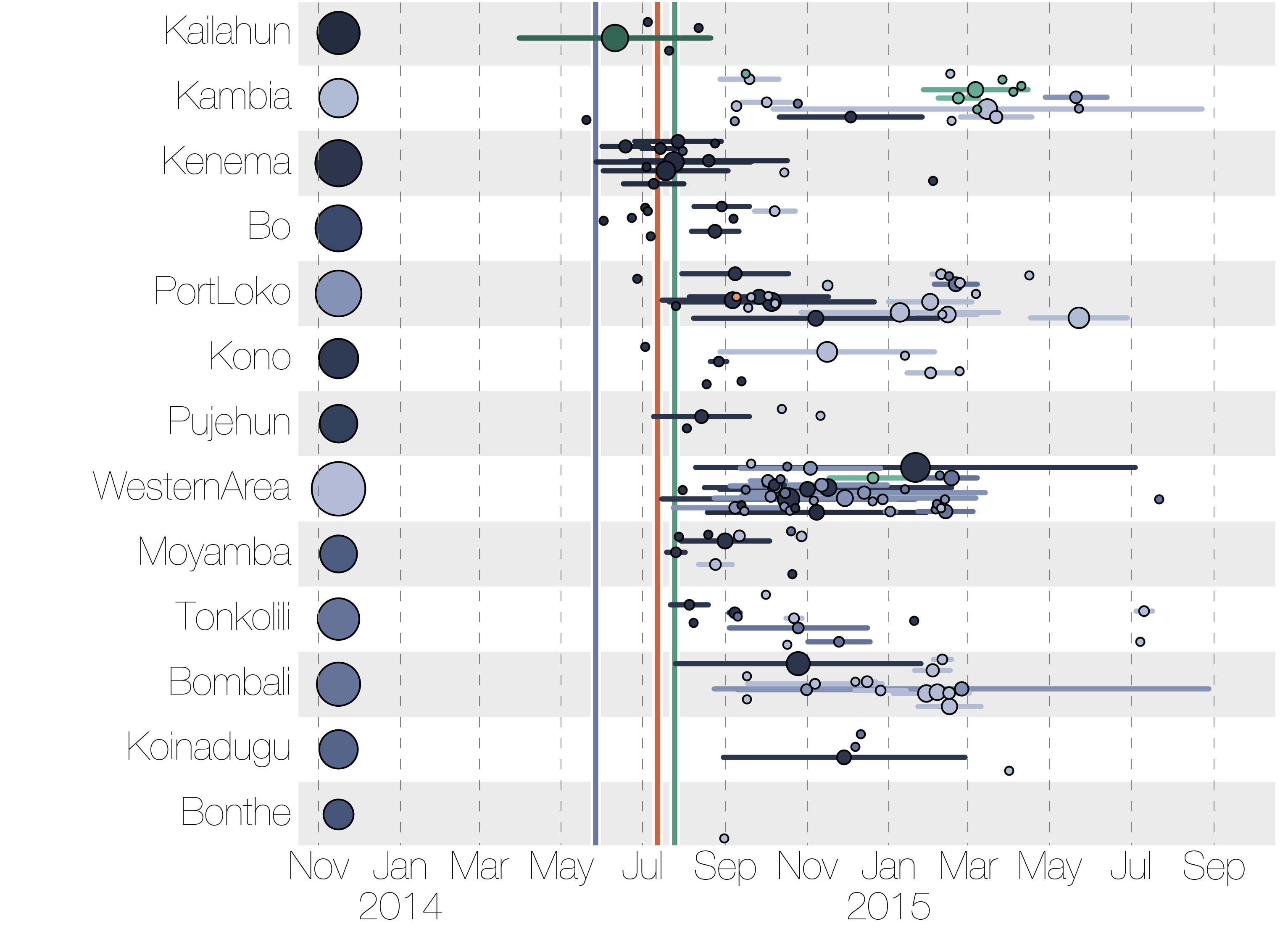
Regional outbreaks due to multiple introductions
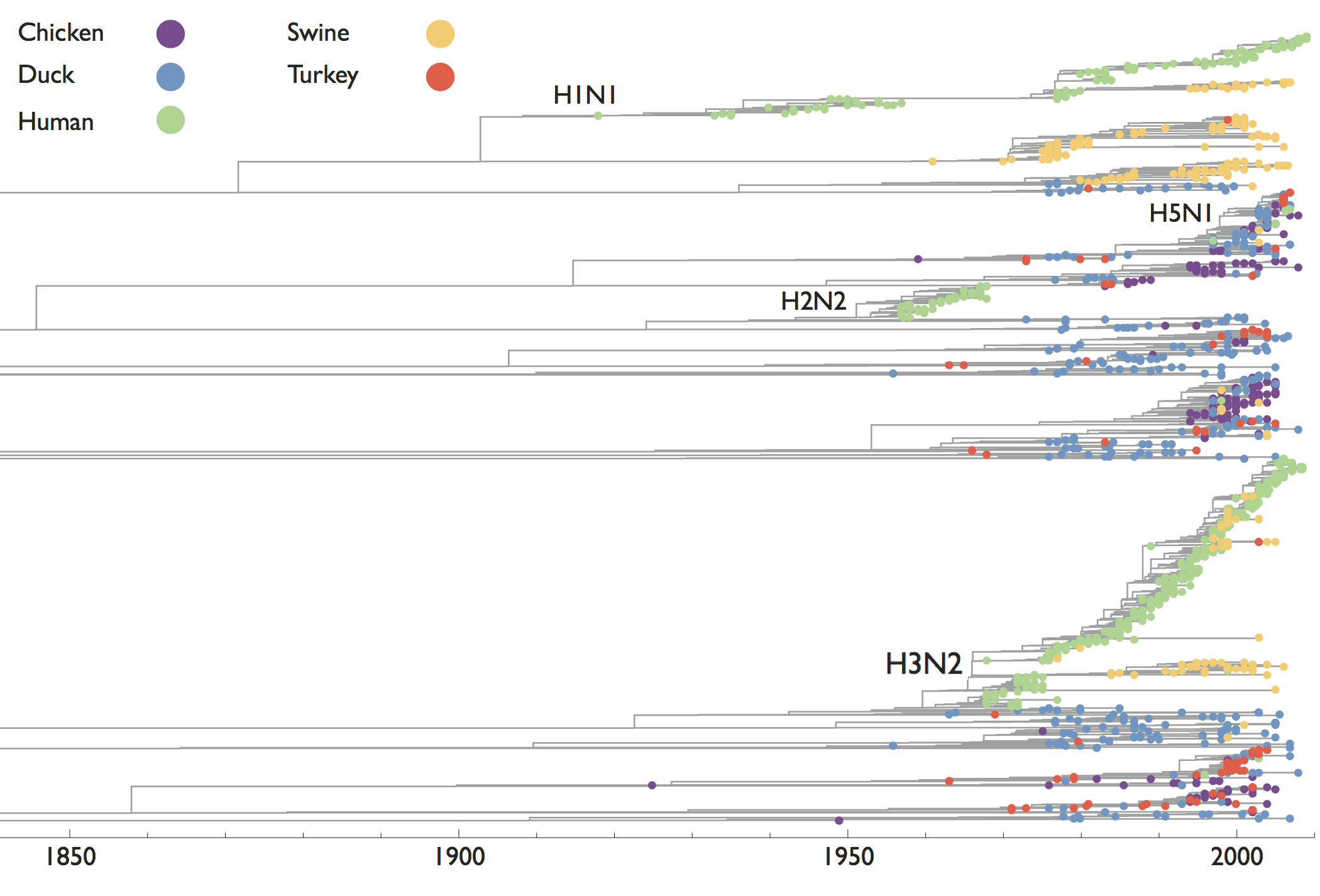
Reservoir species and host jumps
Influenza has a reservoir in wild birds, spillovers sometimes become endemic
Each Ebola outbreak derives from a separate spillover event
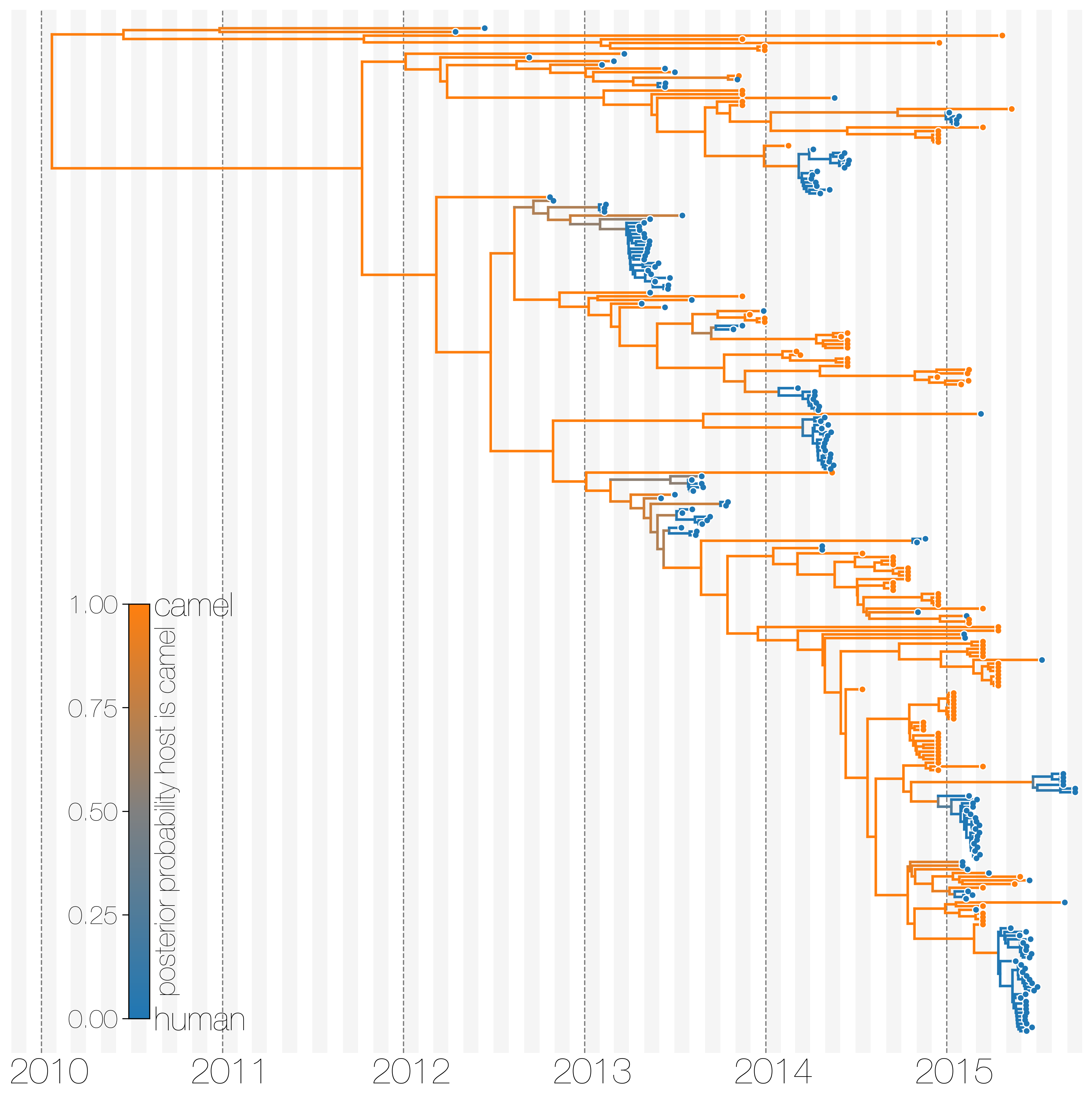
MERS-CoV has frequent spillover events, but limited human-to-human transmission
Reassortment and recombination
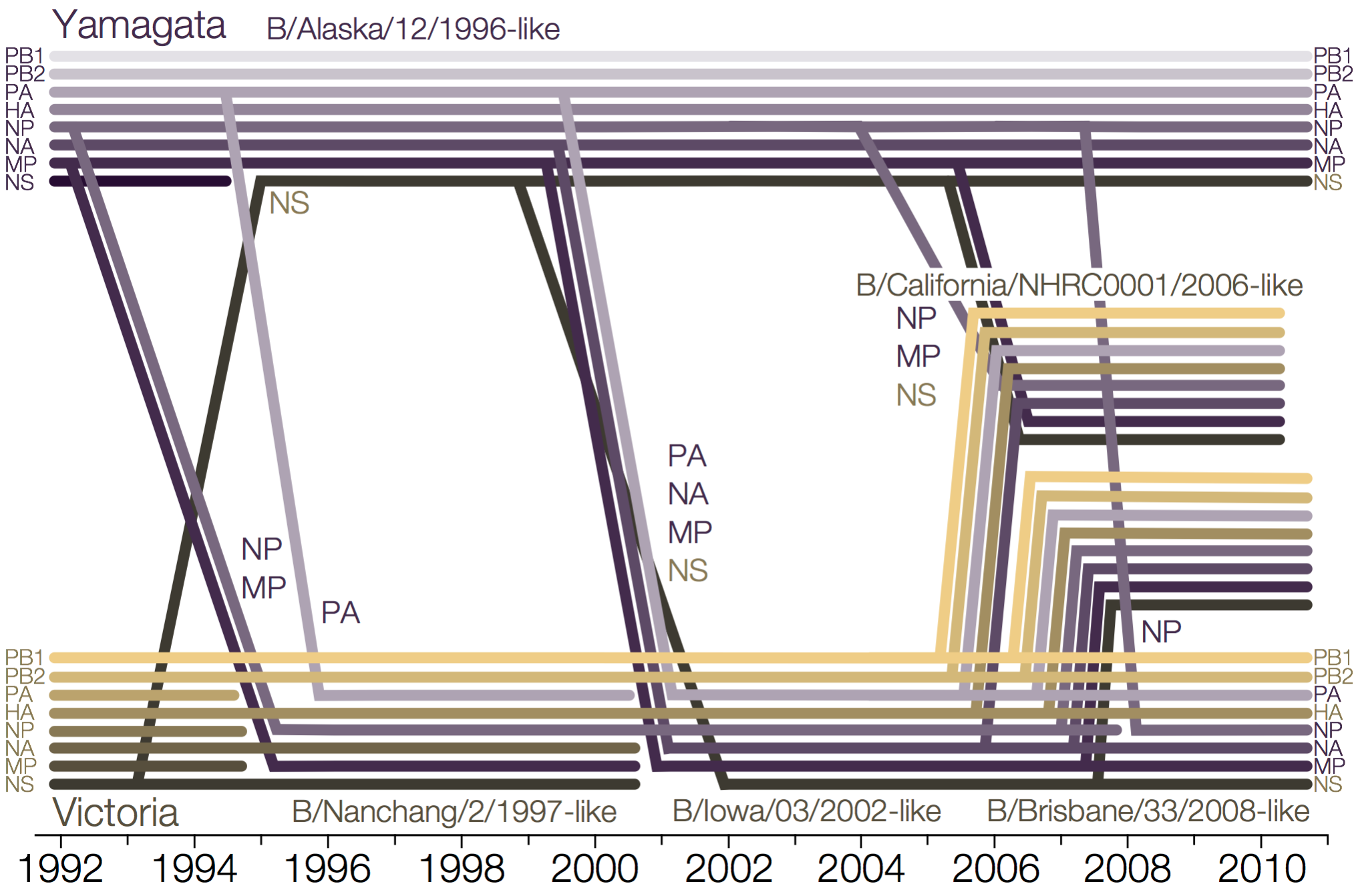
Influenza B reassorts across segments
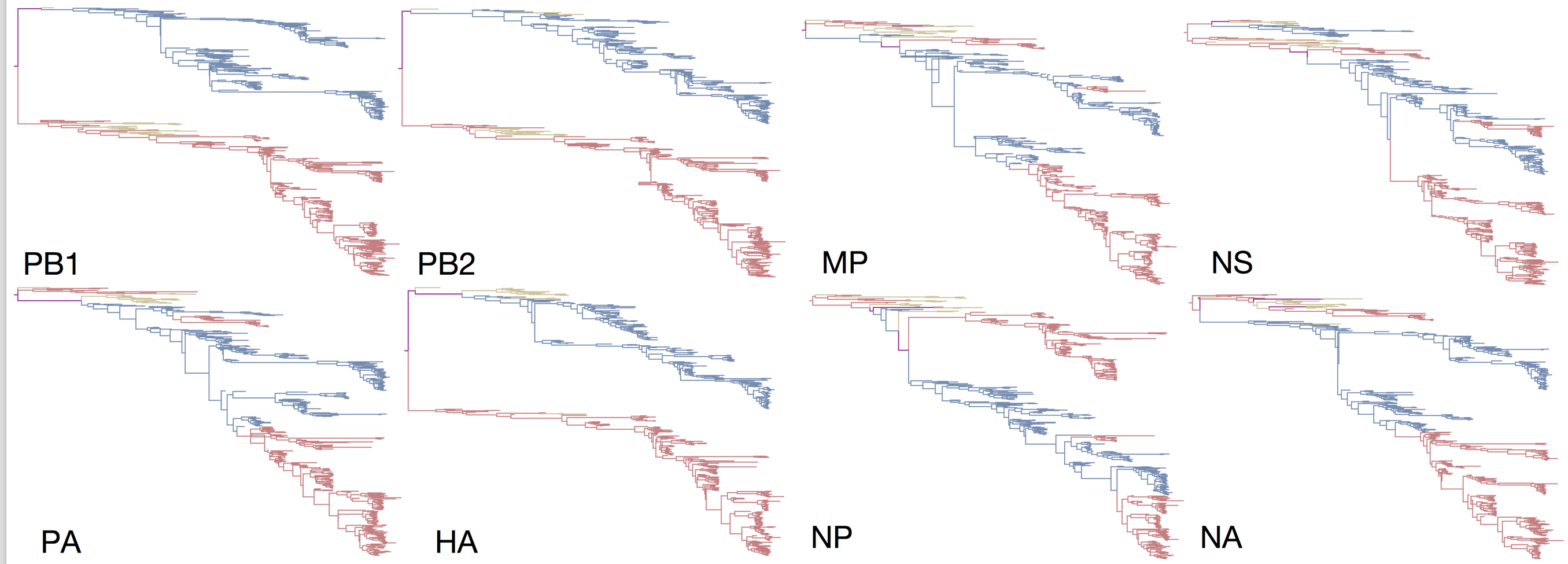
Reassortment splits gene constellations
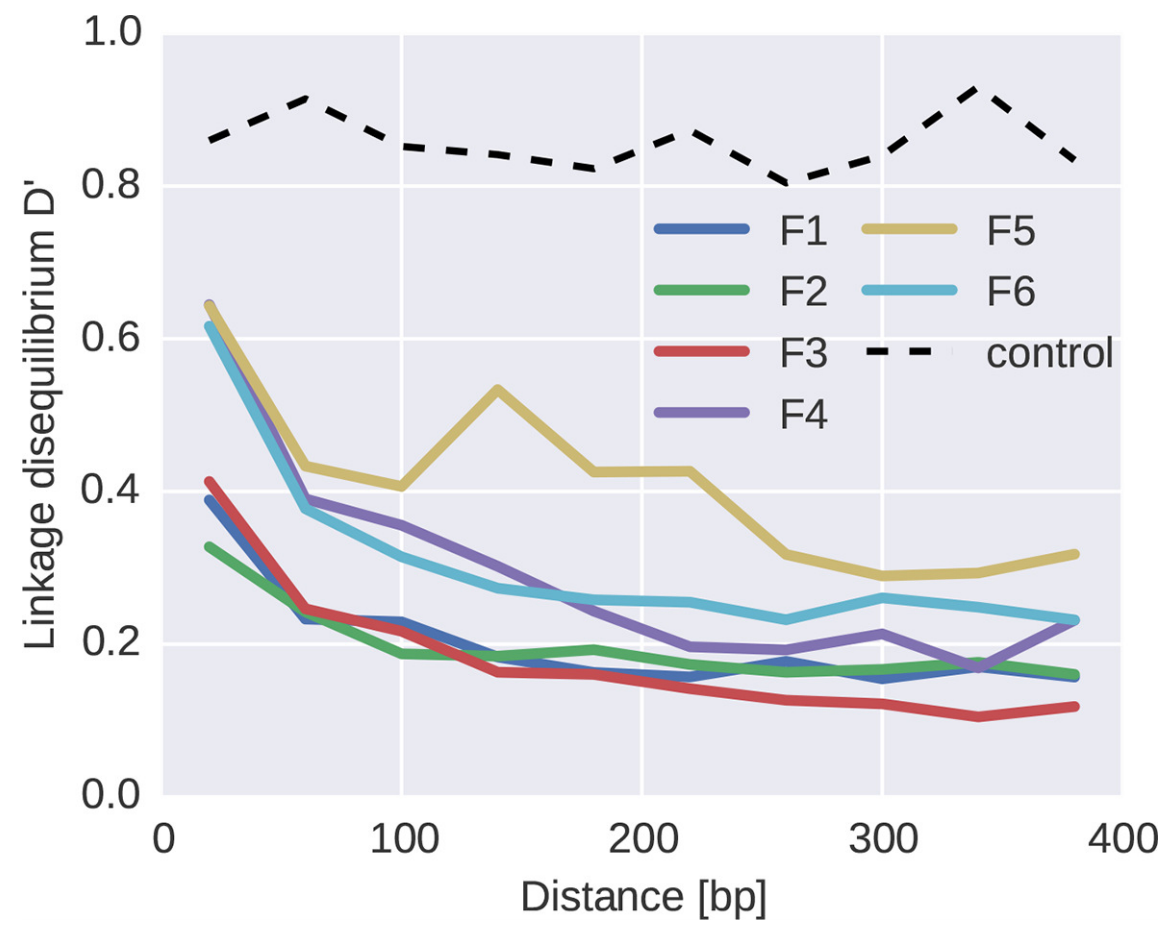
Abundant recombination during within-host HIV evolution
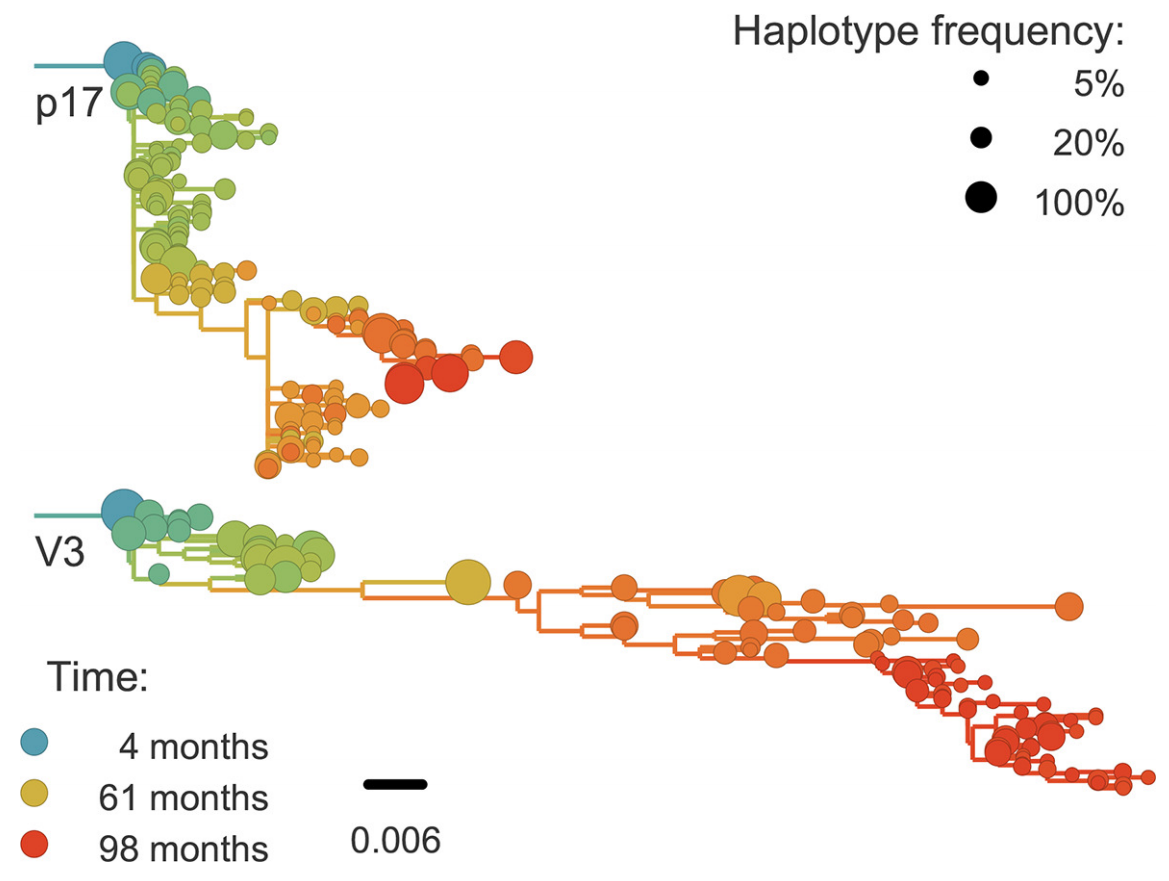
Linkage disequilibrium decays with physical distance